Dispersal and habitat dynamics shape the genetic structure of the Northern chamois in the Alps
Glenn Yannic and Loïc Pellissier shared senior authorship.
Handling Editor: Camila Ribas
Abstract
Aim
Understanding the drivers of species distribution ranges and population genetic structure can help predict species' responses to global change, while mitigating threats to biodiversity through effective conservation measures. Here, we combined species habitat suitability through time with process-based models and genomic data to investigate the role of landscape features and functional connectivity in shaping the population genetic structure of Northern chamois.
Location
European Alps.
Taxon
Northern chamois (Rupicapra rupicapra).
Methods
Using a model that simulates dispersal and tracks the functional connectivity of populations over dynamic landscapes, we modelled the response of the chamois to climate change from the last glaciation (20,000 years ago) to the present. We reconstructed species habitat suitability and landscape connectivity over time and simulated cumulative divergence of populations as a proxy for genetic differentiation. We then compared simulated divergence with the actual population structure of 449 chamois (with >20 k SNPs) sampled across the Alps.
Results
We found that Alpine populations of chamois are structured into two main clades, located in the south-western and the eastern Alps. The contact zone between the two lineages is located near the Rhone valley in Switzerland. Simulations reproduced the geographic differentiation of populations observed in the genomic data, and limited dispersal ability and landscape connectivity co-determined the fit of the simulations to data.
Main conclusions
The contemporary genetic structure of the chamois across the Alps is explained by limited functional connectivity in combination with large rivers or valleys acting as dispersal barriers. The results of our analysis combining simulations with population genomics highlight how biological characteristics, habitat preference and landscapes shape population genetic structure over time and in responses to climate change. We conclude that spatial simulations could be used to improve our understanding of how landscape dynamics, shaped by geological or climatic forces, impact intra- and interspecific diversity.
1 INTRODUCTION
Investigating the effects of historical climatic changes on species distributions, and genetic diversity and structure, can improve our understanding of species demography and help us to predict species' responses to climate changes (Yannic, Pellissier, Ortego, et al., 2014). Because adaptation of large mammals to rapid environmental change is limited and slow (Hetem et al., 2014), their immediate response to climate change is generally a range shift (Chen et al., 2011). The reorganization of species' distributions reshapes the connectivity of populations and, as a consequence, their population genetic structure (hereafter genetic structure), especially in the context of major glacial cycles (Hewitt, 1999; Pellissier et al., 2014). For example, the range of cold-adapted species from higher latitudes, such as the caribou (Rangifer tarandus), shifted southward during glaciations, where they were isolated in multiple refugia, resulting in distinctive signals in their genetic structure (Taylor et al., 2021; Yannic, Pellissier, Le Corre, et al., 2014). Moreover, range shifts depend on the ability of species to disperse to suitable areas over time (Williams & Blois, 2018); consequently, the study of genetic structure can indicate a species' ability to shift its range in interaction with landscape structure (He et al., 2013). Therefore, by combining species habitat preferences over dynamic landscapes with model simulations and genetic data, we can better understand species' past dynamics and identify their capacity to respond to climate change.
As a result of climate or productivity, animal species generally use only a fraction of the landscape (Peterson et al., 1999); when combined with direct barriers to dispersal, such as topography, ice or rivers, these factors determine the connectivity among populations and their genetic differentiation (Frankham et al., 2002). Typically, species in more complex landscapes, such as mountain ranges, show high levels of genetic differentiation between populations (Badgley, 2010). Genetic differentiation can be the result of recent or more ancient habitat fragmentation, such as that associated with distinct glacial refugia (Hewitt, 2004) or the formation of river valleys during geomorphological processes (Hazzi et al., 2018). The combination of landscape dynamics and species dispersal abilities is expected to have modulated the response of species to past environmental change (Hewitt, 1999). For example, genetic analysis has shown that the mountain hare (Lepus timidus) probably recolonized Ireland over a land bridge from southern France (and not from Scotland) following the last glaciation (Hamill et al., 2006), whereas the pygmy shrew (Sorex minutus) was only able to colonize Ireland with anthropogenic translocations from Britain (McDevitt et al., 2011). While historical species dynamics have been inferred from genetic data since the emergence of the field of phylogeography (Hewitt, 1999, 2004), the development of palaeo-environmental reconstructions and species distribution models (Elith & Leathwick, 2009) and process-based models (Landguth & Cushman, 2010) offers new possibilities for insights into species range dynamics (Yannic et al., 2020).
Combining habitat suitability models (HSMs) with process-based simulations enables the reconstruction of species dynamics through geological time and helps us to infer species demographic parameters (Arenas et al., 2012; Yannic, Pellissier, Ortego, et al., 2014), thus representing a central tool for geogenomics. HSMs have been widely used to predict species geographic distributions (Guisan et al., 2017) and to estimate the location of refugia during the glaciations, often in combination with genetic data (Svenning et al., 2011). To be compared with genetic data, however, HSMs must be combined with models capturing actual processes of dispersal and genetic differentiation (Epperson et al., 2010). These process-based models use a bottom-up approach to simulate populations, following rules defined a priori (e.g. for dispersal) to scrutinize emerging patterns (e.g. in genetic structure; Grimm et al., 2005). HSMs have been used as input in process-based models to explicitly simulate the processes of individual dispersal and genetic drift that shape population isolation (Brown & Knowles, 2012). Since individual-based models are computationally demanding (Currat et al., 2019), another class of more parsimonious spatially explicit simulations has been proposed to investigate and validate multiple hypotheses regarding the role of dynamic landscapes in shaping inter- and intraspecific diversity (Leprieur et al., 2016; Yannic et al., 2020). By combining process-based models with species HSMs and exploring biological parameters, simulations can help us to investigate the effects of species dispersal ability (Knowles & Alvarado-Serrano, 2010) and landscape features on genetic structure (Spear et al., 2010). Models can potentially simulate the connectivity of populations over time based on climate reconstructions, and the comparison of these simulations with empirical genetic data can provide insight into past and future species distribution range dynamics (Yannic, Pellissier, Ortego, et al., 2014).
The Northern chamois (Rupicapra rupicapra) is an alpine ungulate native to Europe and the Near East (Corlatti et al., 2011). It has previously been used to study the impacts of habitat connectivity on population genetics (Crestanello et al., 2009; Rodríguez et al., 2008), as its preference for steep, rugged areas (Corlatti et al., 2022) means that its preferred habitat is naturally fragmented (Buzan et al., 2013; Šprem & Buzan, 2016). Also, gene flow is expected to be limited among some populations (Buzan et al., 2013) because wide valley floors may act as natural barriers to dispersal (Soglia et al., 2010). Therefore, simulating population connectivity based on habitat suitability and comparing these models with observed genetic structure could be informative regarding the effect of landscape features on chamois dispersal (Shirk et al., 2012).
In this study, we aimed to provide insight into the drivers of the contemporary distribution and genetic structure of Northern chamois in the Alps through a combination of HSMs, process-based models to simulate cumulative functional connectivity through time, and a comprehensive newly generated genomic data set (>20 k SNPs). We analysed the contemporary genetic structure to determine the geographic distribution of genetic clusters. Next, we simulated the habitat suitability and connectivity from the last glaciation (20,000 years before present, bp) to the present to track functional connectivity. Cumulative distances between clusters that integrate the evolution of connectivity through time were used to generate a divergence matrix, which was compared with the observed genetic structure. We estimated dispersal abilities of the Northern chamois for different landscape feature scenarios by comparing simulations with the genomic data. We investigated the impact of landscape features on population structure in comparison to different dispersal abilities. Based on the expectation that cumulative distances serve as an approximation for population differentiation, we made the following predictions: (1) life history traits such as dispersal can be estimated realistically from simulations; (2) simulations can estimate the relative impacts of landscape features and dispersal on population genetic structure, that is, whether limited dispersal alone is sufficient to explain the observed genetic structure.
2 MATERIALS AND METHODS
2.1 Sampling, ddRADSeq library construction and sequencing
A large network of collaborators (i.e. hunting administrations, non-governmental organizations and National Parks; see Acknowledgements) collected Northern chamois samples (muscle, blood and hair) across the European Alps. From the initial sampling, we excluded samples from Slovakia, as the chamois in the Tatra mountains form an endemic subspecies (R. rupicapra tatrica) and were isolated from other populations for at least 10,000 years (Jamrozy, 2006) before chamois from the Alps were introduced in the last century, leading to hybridization between the two subspecies in the Low Tatra mountains (Zemanová et al., 2015). We used a set of 449 chamois originating from France (n = 115), Austria (n = 110), Italy (n = 95), Switzerland (n = 85), Slovenia (n = 23) and Croatia (n = 21). The samples were genotyped using a double-digest Restriction-site Associated DNA sequencing approach (ddRADSeq; Peterson et al., 2012), and SNP calling was performed using Stacks v2.4 (Catchen et al., 2011, 2013). The detailed procedure regarding ddRADSeq library construction, sequencing and data processing is provided in the Supporting Information. SNPs were filtered out if not genotyped for at least 85% of the samples when the SNP error rate was <2.5% based on the analyses of replicates (~12.5% of the samples, n = 81), and when sequencing read depth was less than 10× or greater than 25×. We only kept samples that were genotyped for at least 75% of the SNPs. The final data set encompassed 20,998 SNPs at the end of the de novo SNP calling procedure. Individuals scored on average 20,102 SNPs ±893 (SD), resulting in 5.7% missing data in the genotype matrix.
2.2 Geographic genomic structure
We analysed the genetic structure of Northern chamois using the ‘adegenet’ R-package v2.1.3 (Jombart, 2008) to infer the optimal number of populations, based on spatial principal component analysis (sPCA) (Jombart et al., 2008) and discriminant analysis of principal components (DAPC; details are provided in the Supporting Information) (Jombart et al., 2010). Additionally, we used Admixture (Alexander et al., 2009) to assess admixture among the populations and determine the optimal number of populations based on the cross-validation error. We computed 10 runs with K ranging from 1 to 8, and we summarized Admixture results using Clumpak (Kopelman et al., 2015). We obtained the genetic distance matrix by calculating pairwise Euclidean distances between individuals (Mastretta-Yanes et al., 2015) based on the SNPs with the ‘adegenet’ R-package. This distance matrix was used for the phylogenetic trees, the Mantel test and the comparison between simulation outputs and empirical genomic data. To assess the population structure, phylogenetic trees were created with hierarchical clustering, applying Ward's distance (Ward Jr, 1963) implemented in R and using the ‘ape’ R-package v.5.4 (Paradis et al., 2004). Additionally, to test for isolation by distance (IBD), we used the ‘ade4’ R-package v.1.7–15 (Dray & Dufour, 2007) to perform a Mantel test (Mantel, 1967). We calculated the pairwise Euclidean geographic distances and the pairwise connectivity distance between each pair of populations using circuit theory (McRae & Beier, 2007) implemented in the ‘gdistance’ R-package v.1.3–1 (van Etten, 2017). Dispersal costs for the connectivity distance were based on the habitat suitability of the best-performing HSM (see next section), similar to the approach of Yannic, Pellissier, Le Corre, et al. (2014).
2.3 Modelling habitat suitability
2.3.1 Occurrences and pseudo-absences
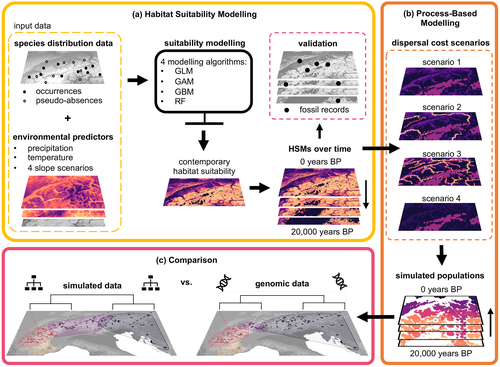
The entire study area encompasses the Alps, the Massif Central, the Jura mountains, the Vosges, the Black Forest and the northernmost part of the Dinaric Alps, spanning from −6° to 19° longitude and from 41° to 51° latitude (see Figure 1 for a general workflow). We collected data on the current species distribution from various citizen science projects throughout the study area, and from hunting records from Slovenia, at a 1 km resolution from the last 15 years (see Table S1). Most of the records of the citizen science data have been validated by experts. We used four different occurrence/pseudo-absence sampling strategies (see also Supporting Information) after experimental testing for more combinations and a wider range of parameters, repeating each of them 10 times (Barbet-Massin et al., 2012). We sampled 10,000 occurrences randomly and 10,000 pseudo-absences either with a slight, middle or strong density dependence with the ‘spatstat’ R-package v1.61–0 (Baddeley & Turner, 2005). Additionally, we applied proportional occurrence sampling combined with random pseudo-absence sampling to account for regions with higher densities of occurrences (Phillips et al., 2009). Details are provided on p. 12 of the Supporting Information.
2.3.2 Environmental variables
After preliminarily testing a set of seven climatic variables that potentially restrict habitat suitability for chamois (cf. Corlatti et al., 2022) for multicollinearity (Guisan et al., 2017), we chose two climatic variables, that is, mean annual precipitation and mean annual 2-m air temperature at 0.008333° resolution from CHELSA-TraCE21k (Karger et al., 2017, 2018) to model habitat suitability through time to the glaciation 20,000 years bp (see section below). To test the extent to which chamois rely on steep terrain, we first modelled the distribution without any terrain variables (only the two climatic variables) and then with four topographic variables derived from the Copernicus Land Monitoring Service (2016): mean slope, slope heterogeneity, a binary classification of suitable slopes for the Northern chamois (20–40°; Bačkor, 2010; Papaioannou et al., 2015) and the number of suitable cells at 100 m resolution within ~900 m grid cells. The environmental predictors showed a correlation of r < 0.7 and a variance inflation factor of <3, values considered acceptable for including all parameters in the same model (Guisan et al., 2017; Zuur et al., 2010).
We used data from Seguinot et al. (2018) for the predicted glaciated areas in the Alps, Jura, Vosges and Black Forest until the glaciation. Additionally, we modelled the glaciers from the Massif Central, estimating the equilibrium line elevation based on the mean annual temperature (Linsbauer et al., 2013) from CHELSA-TraCE21k, and we calculated the area proportional to the glacial extent during the glaciation (Van Vliet-Lanoë et al., 1991).
2.3.3 Habitat suitability
To model habitat suitability, we used four statistical modelling approaches in R v.3.6.1 (R Core Team, 2019), following Yannic et al. (2020) for model creation, validation and prediction at a resolution of 0.0083° back in time until the glaciation in 100-year time steps. We validated the hindcasted habitat suitability with a comprehensive fossil collection using the Boyce index (Boyce et al., 2002). The fossil collection included data provided by C. Callou, personal communications, and the IANUS data portal (Heinrich et al., 2016), and also from literature research (Table S2). A detailed description of the habitat suitability modelling is provided on p. 13 of the Supporting Information.
2.4 Simulating population divergence through time
2.4.1 Simulating populations
We used the process-based model gen3sis from the ‘gen3sis’ R-package v.1.0 (Hagen et al., 2021) to track population functional connectivity through time and to simulate cumulative divergence, based on habitat suitability, dispersal ability and connectivity. This model tracks the number of time steps when two populations are not connected in a pairwise distance matrix rather than simulating the underlying evolutionary processes of population divergence (e.g. drift, mutation and selection). The model assumes that the absence of connectivity increases population genetic differentiation, as has been reported for chamois (Buzan et al., 2013). Given the trade-off due to computational limitations between highly detailed landscapes and genomic processes, this simplification is necessary to explore dispersal traits and complex landscape features through time with thousands of simulations. Gen3sis uses a clustering algorithm to group cells and consists of two main steps: Step 1: two inhabited cells are connected and clustered together if the least-cost path between them is smaller than the dispersal ability of the cells (see section below). Over time, disconnected cells accumulate divergence by a user-defined number per time step, resulting in a cumulative divergence matrix that integrates the effects of barriers to gene flow on population differentiation (Hagen et al., 2021). These divergences are stored in a matrix tracking pairwise divergence of all cells. Connected cells reduce the divergence until it equals 0. Cells exhibiting 0 distance between each other and similar distances to all other cells are assigned to one population. Step 2: the population can disperse to cells which are newly classified as suitable habitat. This requires that the least-cost path to the new cell(s) is within the dispersal ability of at least one cell already inhabited by the population. The divergence is simulated and tracked from the oldest step forward in time. Ultimately, the simulated divergence among groups of cells defines clusters that can be compared with the observed genetic clusters. The selection of suitable cells as input to gen3sis consisted of two steps: first, cells were categorized as suitable if they were classified as suitable by at least two of the four different habitat suitability modelling techniques used. Second, we aggregated the habitat suitability raster to a resolution of 0.05° (~21.4 km2) for the region around the Alps (from 4.6° to 16.3° east and from 43.5° to 48.3° north), given the trade-off between extent, resolution and computational power/storage demand. We classified cells as suitable if more than half of the area of the aggregated cell was suitable at the binary layer at the initial higher resolution (0.0083°). We selected five HSM scenarios (out of 20) to test if chamois distribution is restricted to steep terrain, based on their performance in the past (Boyce index and a visual control of the distribution range) and their fit to the contemporary distribution (TSS and AUC values). We replicated the last time point (20,000 years bp) of the HSMs for 50 time steps as a burn-in to allow for divergence between isolated populations before the dynamic simulation and the recolonization of the Alps started. We assumed that all suitable cells at the oldest time step were inhabited.
2.4.2 Exploring dispersal parameters
We ran simulations with different values for the dispersal parameter to estimate whether the dispersal capacity per time step, which influences both the colonization of vacant cells and the functional connectivity among occupied cells, modulates the fit of the final simulation step to the genomic data (prediction 1). Dispersal ability was chosen randomly from a Weibull distribution for each cell. We explored the parameter settings listed in Table S5 to investigate a range of varying dispersal kernels. The median dispersal ability (d) of the six different scenarios ranged from 0.055 to 0.69 km per year, corresponding to 0.34 to 4.31 km per generation, assuming a generation time of 6.2 years (Gaillard, 1992, as cited in Pérez et al., 2002). This range is within the range of dispersal distances reported by Loison et al. (2008).
2.4.3 Exploring landscape features
To test the impact of landscape features (prediction 2) by combining simulations and genomic data, we considered different costs to cross landscape features. We calculated the cost distances between all cells classified as suitable habitat for Northern chamois using the ‘gdistance’ R-package. This package allowed us to include various dispersal costs of different landscape features, for example, large rivers as dispersal barriers. The dispersal ability describes how far individuals/populations could move on suitable habitat considering the landscape features. As the role of rivers and valleys as dispersal barriers for chamois is debated (Safner et al., 2019; Soglia et al., 2010), we investigated whether rivers indeed form an additional dispersal barrier to chamois, or if the lower habitat suitability of valley bottoms might be enough to form the observed genetic differentiation. Therefore, we created the following four landscape scenarios (see Table S6 for details) to explore their effect on the population structure: in scenario 1, dispersal costs to cross one cell were scaled reciprocally to the habitat suitability (hereafter ‘suitability only’). Scenario 2 was the same as scenario 1 but also included large rivers (e.g. the Rhine) from the Natural Earth (2018) data set as dispersal barriers (‘suitability with barriers’). The costs of crossing rivers decreased with increasing elevation, assuming that valleys and rivers become smaller with increasing elevation and thus are likely easier to cross for chamois (Safner et al., 2019). Scenario 3 was the same as scenario 1 but also included medium (e.g. Aare and Reuss) to large rivers as dispersal barriers from the Food and Agriculture Organization of the United Nations (2014) (‘suitability with medium barriers’). Costs of crossing these rivers increased with their Strahler order, as the Strahler order and thereby the river size increased with each river branching downstream (Safner et al., 2019; Strahler, 1952). In scenario 4, the costs of crossing one cell of unsuitable habitat were set to 1.25 times higher than crossing suitable habitat, irrespective of their suitability. Thus, this scenario depicted mainly geographic distances. Additionally, we tested three different costs of crossing glaciers, ranging from half as expensive to cross, to equal, to twice as expensive to cross than unsuitable land cells. We used the ‘vegan’ R-package v.2.5–7 (Oksanen et al., 2019) to compute the principal coordinate analysis (PCoA) to compare the simulation at different points in time. An overview of all parameters used is presented in Table S7.
2.5 Comparison between genomic data and simulations
We compared the simulated divergence among groups of cells to the empirical genomic clustering. We extracted the most frequent simulated population identity of all sampling locations within a buffer circle of 6 km radius, accounting for dispersal and spatial uncertainty at the raster resolution of ~4.6 km. Knowing the simulated population identity, we used the cumulative divergence matrix simulated by gen3sis to obtain a pairwise distance matrix between all sampling locations. Subsequently, we compared the simulated distance matrix with the actual genetic distance matrix. We excluded samples that were not predicted from the comparison. We calculated the average of two similarity metrics ranging from zero to one to estimate the fit between the simulations and the genomic data. Values closer to one indicate a better fit between the two data sets. First, we calculated a PCoA for the genetic distance matrix and the simulated distance matrix using the ‘ade4’ R-package and compared the two using a Procrustes analysis (Peres-Neto & Jackson, 2001) implemented in the ‘vegan’ R-package. Second, we calculated Cramer's V (Cramér, 2016) to compare the association between the genetic group assignment based on Admixture and the group assignment of the simulation using hierarchical clustering. We fitted GLMs in R with a quasi-binomial distribution between the simulation fit and the parameter settings as explanatory factors. We then used the ‘ecospat’ R-package v.3.1 (Di Cola et al., 2017) to calculate the deviance explained by each single model parameter of the fit between the observed genetic distances and the simulated divergence.
3 RESULTS
3.1 Geographic genomic structure
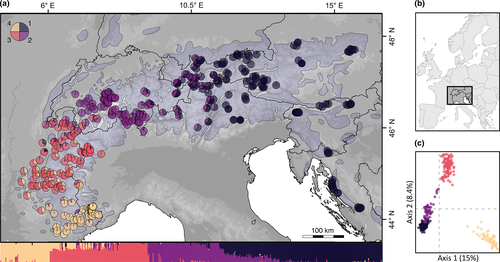
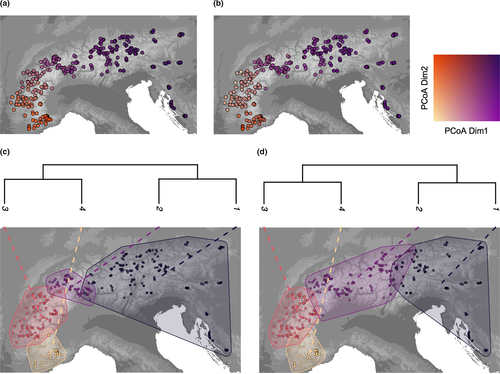
Our SNP analyses showed that the European Alps host two divergent genetic clades of Northern chamois. The eastern part of the Alps including the Dinaric Alps contain one large genetic cluster, whereas multiple clusters occur in the south-western Alps (Figure 2a). The first two axes of the DAPC exhibit a v-shape (Figure 2c), with the chamois from the central and eastern Alps (clusters 1 and 2 in Figure 3) being close to each other and showing some overlap. Consistently, the phylogenetic tree (Figure 3c) showed two main clades, dividing the western and eastern Alps, each with two clusters. We found signals of significant isolation by distance across the Alps according to the Mantel test correlation between pairwise genetic distances and connectivity distance among individuals (Mantel's r: 0.338; p < 0.01). The Admixture plot indicates admixture at the contact zone between different clusters, for example, around the upper Rhone or Adige valleys, but the overall genetic structure was strong in both the Admixture (Figure 2a,b) and multivariate analyses (Figure 3 and Figure S7).
3.2 Predictability of the genetic population structure
The simulation with the best fit to the genomic data according to the similarity metric had a Cramer's V and a Procrustes correlation of 0.88. As with the genomic data, the simulation predicted a main gradient from the south-western Alps to the (north-)eastern Alps (Figure 3). The simulation generally predicted two groups congruent with the observed clades, with the contact zone occurring in the same region as the one derived from the genomic data, that is, the Rhone valley in Switzerland. Each of these main groups is divided into two clusters. The contact zone of clusters 1 and 2 is located further east than the observed one and lies in the Adige valley, whereas the contact zone between clusters 3 and 4 is very similar in the simulation and real data sets. The impact of different parameters on the simulations is presented in the following paragraph, and the results from the habitat suitability modelling are provided in Figure S8 and Tables S3 and S4.
3.3 Dispersal parameters and landscape features
We found that dispersal ability was the most important parameter determining the fit between the simulated and genetic distances, accounting for nearly 40% of the variance in the similarity metric (D2-value: 0.39; see Table S8), while the landscape features explained roughly 10% of the variance (D2-value: 0.11). The simulated divergence fitted the observed genetic structure best when large rivers (or valleys) were added as dispersal barriers to the habitat suitability models (scenario 2; Figures S9 and S10). A short dispersal ability alone or in combination with the habitat suitability resistance surface (scenarios 1 and 4) did not lead to the observed genetic structure. Generally, the model fit decreased with increasing dispersal parameter values, with the largest dispersal distances resulting in a single population across the Alps. Adding smaller barriers in addition to large barriers (scenario 3) decreased the model fit for the lowest dispersal parameter values but increased it otherwise. The shape of the dispersal ability distribution accounted for another 4% of the variance in the model fit to the genomic data. Simulations with higher variability in the dispersal parameter value resulted in less divergence among the cells. The best-performing model exhibited a median dispersal distance of 5.5 km/100 years and a Weibull distribution shape of 1. The binary terrain slope classification and the terrain slope heterogeneity outperformed the other HSMs, explaining less than 1% of the deviance of the model fit in our parameter space. The best fitting model included the binary slope classification.
3.4 Simulated connectivity over time
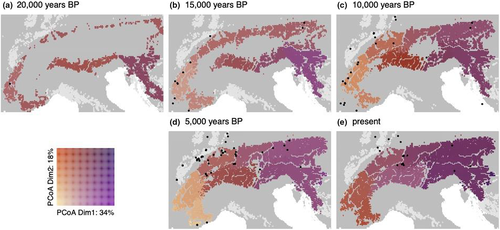
Our simulation predicted that chamois recolonized the Alps mainly from 15,000 years bp onward (Figure 4). The predicted suitable habitat at the south-western and south-eastern part of the Alps during the glaciation was substantially larger than the small and scattered areas at the northern edge. Some fossil records lay outside the focus area (European Alps) and outside of other mountain ranges, but most of them were located in proximity to predicted suitable habitat (Figure 4 and Figure S10). The largest uncolonized area at 10,000 years bp was predicted in the region of the Adige valley, Italy; otherwise, most of the Alps was colonized by then. This predicted long-lasting separation of the populations resulted in the longer separation of clusters east (1) and west (2) of the Adige valley (Figure 3d). The HSM used for the best simulation of the divergence performed well for the present distribution (AUC = 0.90; TSS = 0.67 and Kappa = 0.67; see also Table S3) and less well for the validation of the hindcasted distribution using fossils (Boyce index including buffer: 0.34; Table S4).
4 DISCUSSION
Modelling species' responses to palaeo-environmental changes can help identify the drivers of contemporary distribution and genetic structure (He et al., 2013), and the important features determining spatial and temporal dynamics. The genomic data of 449 Northern chamois samples across the European Alps suggested two main clades: one located in the south-western Alps and one in the eastern Alps. Generally, the model fit based on simulations decreased with increasing dispersal distances, indicating a low dispersal ability of Northern chamois. Additionally, medium to large rivers or valley bottoms acted as important dispersal barriers. Low dispersal ability and habitat suitability alone were not sufficient to depict the observed genetic structure. We showed that the past distribution of species can be reconstructed using dynamic HSMs based on geological and palaeo-environmental data and process-based models. Our approach suggests that combining genomics and palaeo-environmental reconstructions, including data from geology, glaciology or climatology, can elucidate the evolution of intraspecific diversity in complex environments.
Modelling overall population connectivity across dynamic landscapes and comparing the models with genomic data has been shown to reveal the effects of landscape features on genetic structure (Jenkins et al., 2016; Landguth & Cushman, 2010). Our simulation detailed the importance of the complex topographic terrain of the Alps, but also past climate dynamics, in shaping the current genetic differentiation of the chamois. The simulations support previous findings by Soglia et al. (2010) and Zemanová et al. (2015) reporting that large rivers or valley bottoms act as dispersal barriers, although individual chamois can cross rivers (Safner et al., 2019) and on some occasions can even swim in the sea (Kavčić et al., 2020). The barrier to dispersal likely increases with the size and the flow of the river (Safner et al., 2019; Soglia et al., 2010). Rivers can act as natural dispersal barriers for other ungulates such as the bighorn sheep (Ovis canadensis; Deakin et al., 2020) and roe deer (Capreolus capreolus; Coulon et al., 2006). The comparison between the fit of the simulated divergence with and without the terrain slope variable to the observed genetic structure and the validation with fossil records of the different HSM scenarios both confirm that chamois prefer steep terrain, as is commonly accepted (Aellen et al., 1995). It apparently allows them to escape from predators (Bačkor, 2010; Papaioannou et al., 2015) and to avoid competition with other ungulates (Anderwald et al., 2016). Even small steep areas may have been sufficient for the chamois to thrive, since fossil records have been found outside the main mountain ranges, for example, from Belgium (Couturier, 1938) or south-west of the Massif Central (C. Callou, personal communications). Taken together, these results indicate that the comparison of different landscape scenarios in dynamic simulation models can demonstrate the use of complex mountain landscapes by animals and how they shape differentiation among populations.
Dispersal is known to be a key trait influencing species responses to climate change (Schloss et al., 2012). In line with this, we observed that dispersal was the most important parameter determining the fit of the simulation to the genomic data. The best-performing simulations for the Northern chamois suggested a median effective dispersal of 5.5 km/100 years, which is at the lower end of the range observed for chamois in the Alps today (Loison et al., 2008). The better fit of the simulations with a Weibull shape 1 for the dispersal ability distribution reflects the occurrence of many individuals with short dispersal distances and a few with long dispersal, which could reflect philopatric females and male-biased dispersal (Loison et al., 2008), with very few long-distance migrants (Buzan et al., 2013). In fact, in exceptional cases, chamois may disperse up to 60 km (Clarke, 1986), which may reflect the need of individuals to find suitable habitats. Long-dispersing red fox (Vulpes vulpes) individuals, for example, quickly track newly available habitat towards the tundra in response to climate change (Colson et al., 2017; Norén et al., 2015). The simulated dispersal ability was also below that reported for bighorn sheep (Deakin et al., 2020). Since the model only accounted for dispersal resulting from measures in gene flow, it simulated the effective dispersal rate, which is almost always smaller than the real dispersal rate (Kobayashi et al., 2008). Thus, the actual dispersal ability is probably underestimated by our analyses, reducing the predicted habitat that a species facing climate change could use, and affecting management and conservation strategies (Luo et al., 2015).
Our genetic analysis revealed that Northern chamois in the Alps fall into two main groups in the south-west and south-east (Figures 2 and 3; see also Rodríguez et al., 2008). These two lineages probably represent two sources of recolonization of the Alps after the last glaciation, originating from populations isolated from each other during that time. A similar pattern has been shown for red deer (Cervus elaphus; Doan et al., 2022) and the Valais shrew (Sorex antinorii; Yannic et al., 2012). Here, the contact zone of the two chamois lineages lies around the Rhone valley in Switzerland and corresponds to the contact zone found in some alpine plant species that also underwent a range expansion after the last glaciation (Jay et al., 2012), although this appears to be located further west than that of red deer (Doan et al., 2022). However, in contrast to the red deer range expansion following the last glaciation, which was dominated by the north-eastward expansion of populations from south-western refugia (Doan et al., 2022), the recolonization by chamois was likely dominated by the eastern lineage, which expanded west over large parts of the Alps. Another possible contact zone is the Adige Valley; in fact, this valley has been shown to be a barrier during the last glaciation for several alpine mammals, including chamois, but also mountain hare and red deer (Vernesi et al., 2016), separating populations into genetically differentiated clusters east and west of it. The divergence found in the simulation is less pronounced in the genomic data but still visible, for example, as a contact zone with admixed individuals (Figure 2a). The legacy of a long-lasting (>2 million year) separation between the two lineages (Rodríguez et al., 2008) probably overrides the patterns in the genetic structure since the last glaciation to some extent. Additional analysis, involving ancient DNA for example, could provide further insights in the post-glacial history of the chamois. By coupling genomics and simulations, our analyses recovered the likely colonization dynamics of the chamois in the Alps since the last glaciation.
Computational demands limit many modelling approaches, for example, with respect to their resolution (Brown & Knowles, 2012). Although we used a model of simplified evolutionary processes (our counter of divergence was defined entirely by functional connectivity through time), we had to aggregate habitat suitability layers to reduce storage volume and speed up the calculations. The resulting grid cells were about 31–43 times larger than the average individual home range size of the Northern chamois (Nesti et al., 2010). Thus, information on landscape features and connectivity was lost during aggregation, including small suitable habitat patches, resulting in somewhat similar HSM scenarios. This could explain why the topographical variables only marginally affected the model fit. Increasing the spatio-temporal resolution of the gen3sis simulations while simulating longer time spans, for example, until the separation of the chamois lineages, could help to disentangle the drivers of the contemporary population structure. Furthermore, explicitly simulating the action of neutral evolutionary forces (mutation, drift and gene flow) could improve the fit between simulations and genomic data, but would require a reduction in landscape complexity. Other models, such as Splatche3 (Currat et al., 2019) or Nemo (Guillaume & Rougemont, 2006), make fewer simplifying assumptions about evolutionary processes, but are less capable of exploring complex landscape features and dispersal abilities. General limitations of the underlying HSMs of the simulations have already been discussed extensively in the literature (e.g. Elith & Leathwick, 2009), including the issues that arise when hindcasting over millennia (e.g. Svenning et al., 2011). Species interactions may have changed or evolved during the modelled time period, as human overexploitation after the Neolithic at lower elevations likely contributed to the range contraction of the Northern chamois toward alpine areas (Baumann et al., 2005). Thus, part of the ecological niche occupied during or immediately after the glaciation may now be abandoned as unsuitable and not captured when building HSMs based on the contemporary distribution.
5 CONCLUSIONS
Coupling dynamic habitat suitability modelling through space and time, in geologically complex terrain and considering past climate changes, with empirical population genomics improves our understanding of the formation of diversity in complex mountain landscapes. In our case study, a short dispersal distance and specific landscape connectivity dynamics shaped the intraspecific diversity of Northern chamois within its mountain range in the European Alps. Our study demonstrates that such an interdisciplinary and integrative biogeographic approach, linking palaeo-environmental modelling, biological processes (e.g. dispersal) and genomic data, can provide insight into the dynamics of intraspecific diversity. Beyond our case study of one ungulate's response to Quaternary climate change, our approach could be used to study the role of tectonic, geomorphological and palaeoclimatic dynamics in forming intra- and interspecific diversity in mountainous areas (Salles et al., 2019). In the context of far-reaching environmental changes, this framework could be extended to predictions about the future of biodiversity not only in order to inform conservation and population management programs, but also into the past to understand the origin of biodiversity hotspots in mountains.
ACKNOWLEDGEMENT
We thank Philippe Auliac, Aurélie Barboiron, Bruno Bassano, François Biollaz, Glauco Camenisch, Marie Canut, Jérôme Cavaillès, Fabian Fopp, Mathieu Garel, Veronika Grünschachner-Berger, Marie Heuret, Ludovic Imberdis, Hannes Jenny, Martina Just, Christine Lettl, Laura Martinelli, Radka Poláková, Elias Pesenti, Davide Righetti, Christine Saint-Andrieux, Federico Tettamanti, Roberto Viganò, the Office of Dolomiti Bellunesi National Park, the Corpo di Polizia Provinciale di Belluno, the Associazione Cacciatori Trentini, the hunting authorities of Canton Berne and IVB Genetic Bank for helping to collect and/or providing the tissue samples. No additional permits were required for the collection of the tissue samples of the hunted animals. Thibaut Capblancq helped with the bioinformatics. We thank the following people for providing observational data which enabled us to perform HSMs: Andrej Rot, Prof. Anna Loy, Roberto Lardelli, Phillipe Jourde, Stefan Munzinger and Simon Capt. The research benefited from the support of AnaBM (USMB) and AEEM (UGA) laboratory facilities and we are grateful to the Roscoff Bioinformatics platform ABiMS (http://abims.sb-roscoff.fr). We thank Benjamin Flück and Oskar Hagen for their technical support to run gen3sis. We are grateful to Prof. Jörg Schibler for providing fossil records from his study from Switzerland and to Cécile Callou (Muséum National d'Histoire Naturelle, Paris) from France. We thank Julien Seguinot for providing the glacier data. Finally, we acknowledge two anonymous reviewers, Alex Widmer, Oskar Hagen, Melissa Dawes, Tobias Roth and Seraina Leugger for providing comments which greatly improved the quality of the manuscript. LP was supported by the Swiss National Science Foundation grant (N° 310030_188550). Open Access Funding provided by Eidgenossische Technische Hochschule Zurich. [Correction statement added on 18 May 2022, after first online publication: CSAL funding statement has been added.]
CONFLICT OF INTEREST
None declared.
Open Research
DATA AVAILABILITY STATEMENT
The data used in the article can be found at: (1) Genomic data: Fastq files are available under the NCBI BioProject “PRJNA813419” (Accession number: BioSample: SAMN26501511-SAMN26502071 and SRA: SRR18252263-SRR18252823). (2) Environmental data: (I) climate data: https://chelsa-climate.org/downloads/; (II) slope: https://land.copernicus.eu/imagery-in-situ/eu-dem/eu-dem-v1-0-and-derived-products/slope?tab=download. (3) Chamois occurrence data cannot be shared publicly due to the regulations of the providers. Publicly available data can be found at https://www.gbif.org/species/5220170. (4) Chamois fossil records, HSM raster and scripts to run gen3sis are available through EnviDat: https://doi.org/10.16904/envidat.291
REFERENCES
BIOSKETCH
Flurin Leugger is interested in how landscape history shapes today's biodiversity and genetic patterns. He and the other authors collaborate in monitoring biodiversity, developing models to understand the drivers of biodiversity patterns, and forecasting ecosystem responses to global changes (see https://ele.ethz.ch).
Author contributions: GY and LP conceived the ideas and supervised FL. EB, LC, BC, BR, HCH, NS and RV contributed to the sample collection. DR, NCGG, NT and ST performed laboratory work. TB and GY performed the bioinformatic analysis. DNK provided the climatic data. FL compiled the occurrence data, performed the models and simulations, and ran the analysis. FL, GY and LP wrote the first version of the manuscript with the support of all co-authors.




