Histological and Ultrastructural Observation Reveals Significant Cellular Differences between Agrobacterium Transformed Embryogenic and Non-embryogenic Calli of Cotton
Supported by the State Key Basic Research and Development Plan of China (2004CB117305).
Abstract
Over the past few decades genetic engineering has been applied to improve cotton breeding. Agrobacterium medicated transformation is nowadays widely used as an efficient approach to introduce exogenous genes into cotton for genetically modified organisms. However, it still needs to be improved for better transformation efficiency and higher embryogenic callus induction ratios. To research further the difference of mechanisms for morphogenesis between embryogenic callus and non-embryogenic callus, we carried out a systematical study on the histological and cellular ultrastructure of Agrobacterium transformed calli. Results showed that the embryogenic callus developed nodule-like structures, which were formed by small, tightly packed, hemispherical cells. The surface of some embryogenic callus was covered with a fibrilar-like structure named extracellular matrix. The cells of embryogenic calli had similar morphological characteristics. Organelles of embryogenic callus cells were located near the nucleus, and chloroplasts degraded to proplastid-like structures with some starch grains. In contrast, the non-embryogenic calli were covered by oval or sphere cells or small clusters of cells. It was observed that cells had vacuolation of cytoplasm and plastids with a well organized endomembrane system. This study aims to understand the mechanisms of embryogenic callus morphogenesis and to improve the efficiency of cotton transformation in future.
Conventional breeding is an effective tool for cotton improvement. For many desirable traits, however, it is limited to narrow genetic resources. Genetic engineering, facilitated by tissue culture technology, provides an attractive approach to break such restrictions (Wilkins et al. 2000). In the last few decades, there has been progressive success in cotton regeneration and transformation (Wilkins et al. 2000). To efficiently integrate transgenes into the cotton genome, Agrobacterium-mediated transformation is by far the most common method (Sunilkumar and Rathore 2001; Mishra et al. 2003). Although this transformation system is generally more efficient than other approaches, it also requires an efficient plant regeneration procedure. It is more difficult to obtain somatic embryos and regenerated plants from cotton than other crops, such as Oryza sativa L. ssp. japonica and Brassica campestris L. (Trolinder and Chen 1989; Wilkins et al. 2000).
Since Davidonis and Hamilton (1983) first reported cotton regeneration and with several decades of development, numerous cotton regeneration systems and transformation systems have been established from cotton hypocotyl and embryogenic callus (Firoozabady et al. 1987; Zapata et al. 1999; Sakhanokho et al. 2001, 2004; Liu et al. 2004; Leelavathi et al. 2004; Wu et al. 2005; Jin et al. 2006). Although the Agrobacterium-mediated transformation method has significantly improved efficiencies of cotton transformation in recent years, only a limited number of elite cotton varieties can be transformed by this method to obtain transgenic plants. Genotype dependence of this technology still restricts the application of biotechnology on cotton development and fundamental research (Wilkins et al. 2000).
One underlying factor for these obstacles was the prolonged procedure of transformation, causing a decreased regeneration ability of explants and difficulties in embryogenic callus induction (Wilkins et al. 2000). Embryogenic callus induction and potency of embryogenesis are crucial aspects that need to be improved during cotton transformation.
Some research groups conducted light and electron microscopy to study structural and cellular changes involved during acquisition of embryogenic competence. These research reports provided detailed descriptions of the morphological and ultrastructure changes that characteristics of acquisition of embryogenic competence and embryoid initiation (Sunilkumar and Rathore 2001; Poljuha et al. 2003; Moghaddam and Taha 2005; Namasivayam et al. 2006; Mikula et al. 2007; Orban et al. 2007).
In cotton transformation research, more attention has been paid to the optimizing of conditions for establishing transformation system (Liu et al. 2004; Leelavathi et al. 2004; Sakhanokho et al. 2004; Wu et al. 2005; Jin et al. 2006) and there are relatively few published reports related to ultrastructural changes caused by transformation.
To better understand the histological and cellular features of embryogenic callus of cotton, which will ultimately help to develop more efficient methods to induce embryogenic callus formation, we studied histological structure, surface cell organization and ultrastructure of embryogenic and non-embryogenic callus during the course of Agrobacterium-mediated cotton genetic transformation. We analyzed embryogenic callus induction and the potential of embryogenesis of transgenic callus, in which a foreign green fluorescent protein (GFP) gene served as a reporter to facilitate microscopic evaluation (Chiu et al. 1996; Sunilkumar et al. 2002), and chloroplast served as markers of cell differentiation and dedifferentiation.
Results
Callus induction and plantlets regeneration
Hypocotyl segments (Figure 1A) began to swell after they were cultured in callus induced medium (CIM) for 1–2 weeks, and generated a mass of calli 4–6 weeks later (Figure 1B). A total of 50 mg/L kanamycin was supplemented to CIM and 34.5% hypocotyl segments introduced kanamycin-resistance calli. The color of most calli was grayish-yellow and the texture was loose and friable. Such calli could be induced into embryogenic calli easily (Figure 1B). In contrast, a few calli showed white or greenish color and hard texture, and they were difficult to form embryogenic calli. Via the fluorescence microscopy method, 97.0% of kanamycin-resistance calli expressed GFP (Figure 2A). Positive transgenic calli, which were confirmed by polymerase chain reaction (PCR) and Southern blotting were further subcultured and studied.
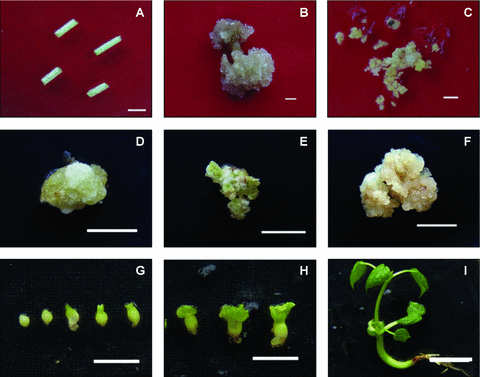
Hypocotyl fragments, calli, somatic embryos, and regenerated plantlets.(A) Hypocotyl fragments (bar, 0.5 cm). (B) Calli originated from edges of hypocotyl explants (bar, 1.0 cm). (C) Embryogenic calli (bar, 0.5 cm). (D) Callus of white color with hard texture (bar, 0.5 cm). (E) Callus of greenish color with hard texture (bar, 0.5 cm). (F) Overgrown callus shows gray-white color and soft texture (bar, 0.5 cm). (G,H) Globular embryos, torpedo and cotyledonary embryos (bar, 0.5 cm). (I) Regenerated plantlet (bar, 1.0 cm).
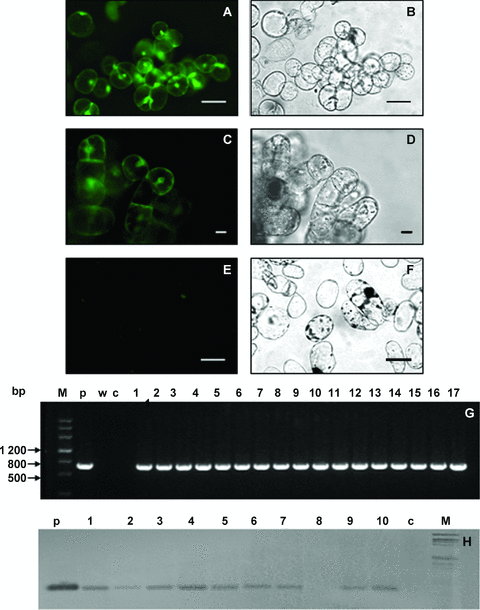
Green fluorescent protein (GFP) expression and genomic DNA analysis of GFP positive lines.(A) Fluorescence image of an embryogenic callus. (B,D,F) Bright field images of each of the calli shown in panels A, C and E (bars, 20.0 μm). (C) Fluorescence image of a white, greenish, hard texture callus. (E) A non-transformed callus. (G) Polymerase chain reaction (PCR) analysis of genomic DNA for the GFP coding region. Lanes: M, DNA molecular weight markers; c, untransformed control plant; p, GFP positive control fragment from pBI-GFP; w, a negative control PCR reaction with deionized water as template, 1–17, GFP positive callus lines. (H) Southern blot analysis of GFP positive lines. p, product of plasmid pBI-GFP digested by BamH I and Nco I; c, Non-transformed plant; M, Hind III and EcoR I digested λ-DNA as a molecular marker.
Grayish-yellow kanamycin-resistant calli that expressed GFP were transferred to solid embryogenic callus induction medium (SECIM) for embryogenic callus induction. Based on the criteria of color, texture, dispersiveness in water, and size/shape of undifferentiated cells (Zhang et al. 2001; Kumria et al. 2003; Mishra et al. 2003), embryogenesis potency of different calli were evaluated. After 6–8 weeks, embryogenic calli were obtained, which showed yellow-green color and nodular friable texture (Figure 1C); in the meantime, different types of non-embryogenic calli were introduced (Figure 1D–F). These non-embryogenic calli showed various colors and textures. Snowy calli appeared to be white and hard in texture (Figure 1D). Greenish calli exhibited yellowish-green and hard texture (Figure 1E). Overgrown calli displayed white to light-brown and soft texture and they dispersed in water easily (Figure 1F).
After embryogenic calli were subcultured in SECIM for 6–8 weeks, globular, torpedo and cotyledonary embryos (Figure 1G,H) appeared. Then, somatic embryos were transferred into somatic embryo germinating medium (SEGM). After being subcultured several times, numerous true leaves grew out, and plantlets were grafted onto rootstock seedlings in soil pots.
Cell organization pattern on surface of embryogenic callus and non-embryogenic callus
Calli formed in cotton transformation varied greatly in texture and color. We observed the cell organization patterns of embryogenic and non-embryogenic calli by scanning electron microscopy.
Embryogenic calli appeared as nodule-like structures, which were composed of small and tightly packed cells. These structures represented a complex of densely packed cells and showed clear boundaries. These cells were hemispherical, small, had similarities in morphology and no wrinkles (Figure 3A). Cells of some embryogenic callus on the surface were connected and covered by the extracellular matrix (Figure 3B).
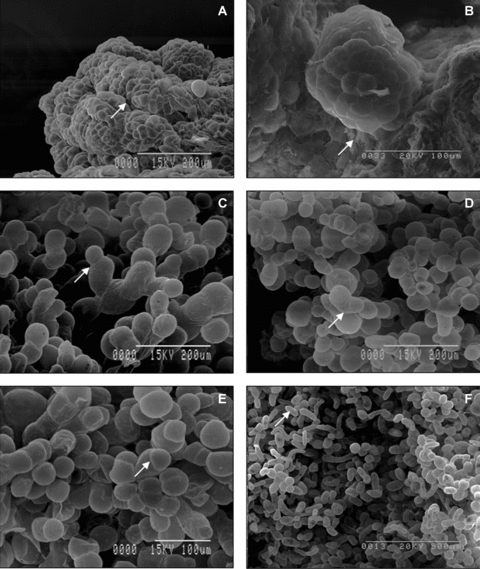
Cell organization pattern on surface of embryogenic and non-embryogenic calli.(A) Densely packed cells on the surface of embryogenic callus represented a series of clusters with clear boundaries (arrow). These cells were smooth, hemispherical and uniform in morphology. (B) Some embryogenic calli presented extracellular matrix surface network (arrow). (C) Cell morphology on the surface of white-hard callus varied greatly; cells divided transversely and formed filament structures (arrow). (D,E) Green-hard callus appeared to have a loose surface structure; cells displayed heterogeneous morphology (arrow) and nodular structures were rare. (F) Most cells on the surface of overgrown callus were arranged singly (arrow).
Non-embryogenic calli were characterized by a high variant morphology of cell organization on the surface. These calli were covered with isolated oval cells or small cell clusters. Results of microscopic observations revealed that there were two distinct types of cells on the surface of calli: large, loosely adjoined oval cells and spherical cells. Meanwhile, cell sizes on the surface of white-hard calli varied greatly (Figure 3C). These cells proliferated transversely and formed filament structures. One such typical structure is shown in Figure 3D,E. Green-hard calli displayed loose surface structures and various cell sizes on the surface (Figure 3E). Cells on the surface of overgrown calli were arranged loosely (Figure 3F, arrow), and filamentous cell morphology suggested that division of these cells was not isotropical (Figure 3F).
Histology and ultrastructure of embryogenic and non-embryogenic calli
In order to study the histological and ultrastructural differences between embryogenic and non-embryogenic calli, histological and ultra-thin sections were observed under a light microscope and an electron microscope, respectively.
Results of the histological observation revealed that early embryogenic callus displayed similar cellular morphology and lack of intercellular spaces (Figure 4A,B; Figure S1A), as well as some nodular structures on the surface (Figure 4A). Some calli formed the embryogenic nodular structures. Compared with the cells of callus, cells that formed embryogenic nodular structures contained dense cytoplasm and appeared to be similar to meristematic cells (Figure 4C). On the cytological ultrastructure, cells of embryogenic calli contained clusters of small vacuoles and had a high electron dynastical cytoplasm. Nuclei and other organelles migrated toward the centre of the cell (Figure 5A). Chloroplasts were degraded to proplastids with some starch grains in which the endometrial system was disassembled (Figure 5E).
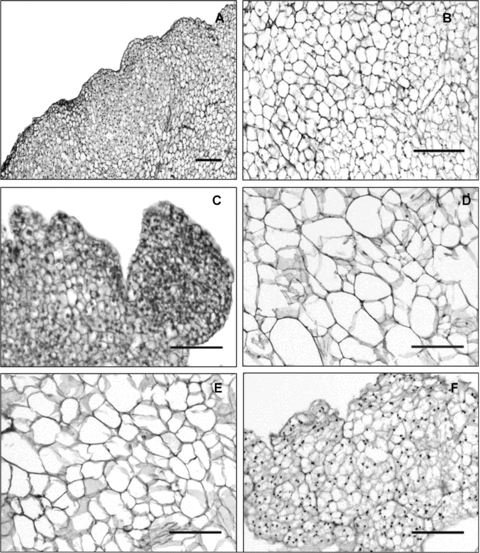
Histological structures of embryogenic and non-embryogenic calli.(A) Histological structure of embryogenic callus. (B) Microscopic image of embryogenic callu core. (C) A detailed histological image of embryogenic nodular structures. (D,E) Histological image of white-hard callus and a green-hard callus, respectively. (F) Micrograph of overgrown callus (bars, 50.0 μm).
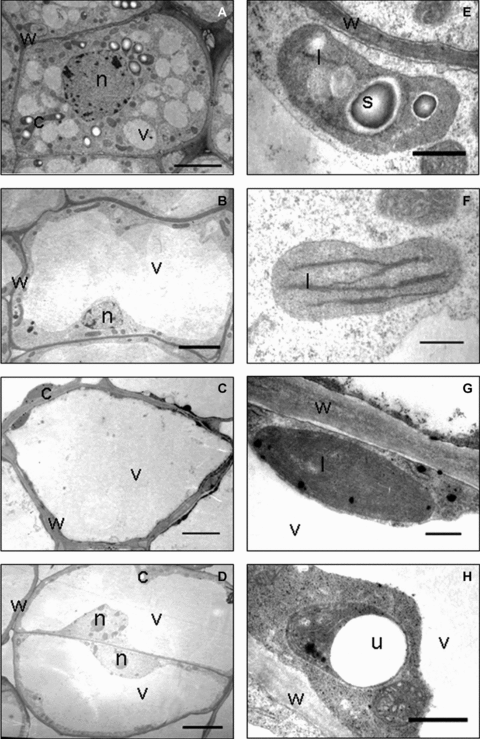
Ultrastructure of different embryogenic calli and non-embryogenic calli.(A–D) Cellular ultrastructure of embryogenic callus cells (A, bar, 5.0 μm), white-hard callus cells (B, bar, 10.0 μm), green-hard callus cells (C, bar, 10.0 μm) and overgrown callus cells (D, bar, 6.0 μm). (E) Morphology of proplastid and degenerated endomembrane system of an embryogenic callus cell (bar, 5.0 μm). (F) A chloroplast of white-hard callus cell contained degenerated endomembrane system (bar, 500.0 nm). (G), A chloroplast of green-hard callus cell contained well-organized endomembrane system (bar, 500.0 nm). (H) The proplastid of overgrown callus cell showed degraded endomembrane system and vesicle-like structure (bar, 500.0 nm). c, chloroplast; l, grana lamella; n, nucleus; s, starch; v, vacuole; w, cell wall.
White-hard calli and green-hard calli had similar histological structures. Shape and sizes of cells, which was composed of these tissues varied greatly. Compared with the loose morphology outside of calli, cells inside calli were compact relatively and developed some intercellular spaces (Figure S1B,C,D). These two types of calli also showed similar features in ultrastructure. The nucleus was located in a narrow strip of cytoplasm between the cell wall and the large vacuole, which is located in the center (Figure 5B,C). Chloroplasts of green-hard calli developed a well organized membrane system, as well as some vesicles and plastoglobuli, which presented in the thylakoids system (Figure 5F). Chloroplasts of white-hard calli showed a similar endomembrane system (Figure 5G). Another type of non-embryogenic calli, the overgrown calli, grew rapidly and was difficult to differentiate into somatic embryos. Results of the histological study showed that cells composed of overgrown calli had similar morphology and a structure of a series of cell clusters could be observed (Figure 4F). Results of the ultrastructure study showed that cells were vacuolized and the nucleus with visible nucleolus was located near the cell wall (Figure 5D). Chloroplasts were mostly degraded into proplastids (Figure 5H). In contrast to the accumulation of starch grains, proplastids in overgrown calli exhibited vacuole-like structures (Figure 5H).
Discussion
Due to the low efficiency of embryogenic callus induction, the Agrobacterium-mediated cotton transformation method is time-consuming and labor intensive. It is believed that calli have different abilities for somatic embryogenesis as their cells are in different conditions and have different characteristics (Pierik 1987). Embryogenic and non-embryogenic calli show differences not only in histological structures and embryogenesis behaviors, but also in their cellular features.
To achieve transgenic cotton lines successfully, the first task is to obtain positive transformed calli for embryogenic callus induction and further studies. In our experiments, 93.2% of loose and friable kanamycin-resistance callus expressed GFP (Figure 3A,B). Such results were confirmed by PCR and Southern blot hybridization analysis (Figure 3G,H). Differences between embryogenic and non-embryogenic calli were then studied systematically, such as the pattern of cell arrangement on callus surface, internal histological structure, as well as the cytological ultrastructure.
Cell arrangement on the callus surface showed significant differences between embryogenic and non-embryogenic calli. Embryogenic calli displayed nodular structures on the surface that were composed of small, tightly packed hemispherical cells, and some calli were covered by extracellular matrix. Although non-embryogenic callus varied greatly in shape, color and texture, they showed similar arrangement of cells on the surface. Non-embryogenic calli were covered by dispersed oval cells or small cell clusters.
The most distinctive character of non-embryogenic callus was the varied cell organizing patterns on the callus surface. Such differences were also described in the somatic embryogenesis of Mammillaria gracillis (Poljuha et al. 2003), wheat (Konieczny et al. 2005), cactus (Konieczny et al. 2005), and barley (Caredda et al. 1999). It appears to be important for plant regeneration that the callus surface is coated by an extracellular matrix network, which appears during the early stages of cotton embryogenesis exclusively (Figure 4B). A similar phenomenon was found in somatic embryogenesis of other plants (Iwai et al. 1999; Verdeil et al. 2001; Konieczny et al. 2005, Popielarska-Konieczna et al. 2008).
At the histological level, another distinct difference between the two tissues was that the embryogenic callus cells showed similar morphology, whereas the hard texture non-embryogenic callus cells appeared very different in cell size and shape; but overgrown non-embryogenic callus appeared similar to embryogenic callus in cell morphology. For ultrastructure, chloroplasts of embryogenic calli cells degenerated to proplastid-like structures with starch grain accumulation. In contrast, chloroplasts of non-embryogenic callus cells showed a well organized thylakoid system. The chloroplasts of non-embryogenic callus were similar to chloroplasts of mesophyll cells. A similar phenomenon was observed in somatic embryogenesis of other plants (Wang et al. 1998; Caredda et al. 1999; Mikula et al. 2007; Orban et al. 2007).
Overgrown callus is a type of non-embryogenic callus, which was characterized by its rapid growing. Cells of overgrown calli showed similar ultrastructures to the embryogenic callus cells. But since they grew rapidly, these cells displayed some cancer cell like characters, such as the appearance of a large number of mitochondria and partially developed cell walls (Astarita and Guerra 1998; Moghaddam and Taha 2005).
Obvious differences in cell organization pattern on callus surface, histological features and cell ultrastructure suggested different potency of embryogenesis between embryogenic callus cells and non-embryogenic callus cells. Although both types of calli developed in the same medium under identical environments, the chloroplasts of embryogenic callus cells showed distinct ultrastructure from cells of non-embryogenic callus. Thus chloroplasts could be used as a cellular marker to estimate totipotency of cotton calli cells.
Differences in cell organization pattern, histological and cytological ultrastructure suggested changes in cell fate. Cascades of signals are involved and cotton somatic embryogenesis may require multiple signaling. Future work should be carried out to understand the signaling pathway involved in introducing embryogenic callus and why non-embryogenic callus lost potency of embryogenesis.
Materials and Methods
Bacterial strains and plasmids
The coding region of an enhanced green fluorescent protein gene (EGFP) was subcloned by BamH I and NcoI restriction enzymes from vector pGJ280 to vector pBI121. The binary vector pBI-EGFP (Figure 6) was then transferred into an Agrobacterium strain LBA4404 by electroporation.

Schematic representation of T-DNA region in the binary vector pBI-GFP. The binary vector pBI-GFP harbors an npt-II gene as a selection marker and a GFP gene as a reporter gene (LB, T-DNA left border; RB, T-DNA right border). EGFP, enhanced green fluorescent protein; NOS, nopaline synthase; NPT, neomycin phosphotransferase II.
Plant materials and growth condition
Seeds of Cotton (Gossypium hirsutum L.) variety CCRI24 were de-coated and sterilized with 0.1% (m/v) HgCl2 solution for 3–5 min and rinsed four times with sterilized deionized water. Aseptic seeds were placed on SGM (seeds germination medium, Murashige and Skoog (MS) basal medium, and solidified with 3.0% (m/v) agar) for germination. Seeds were cultured at 28 °C with a 16-h photoperiod daily.
Cotton transformation
Hypocotyls were excised from aseptic seedlings and cut into 5–7 mm (Figure 1A) segments. The hypocotyl segments were infected by Agrobacterium tumefaciens LBA4404 containing pBI-EGFP in the bacterial suspension (OD600= 0.3) for 3–5 min. Infected hypocotyls were blotted on sterile filter paper to remove excess A. tumefaciens, and then, hypocotyl segments were transferred on a piece of filter paper on the co-cultivation medium (CCM). CCM was made of modified MS salts and B5 vitamins supplemented with 0.05 mg/L indole acetic acid (IAA), 0.05 mg/L kinetin (KT), 0.05 mg/L 2, 4-Dichlorophenoxyacetic acid (2, 4-D), 25.0 g/L glucose, and solidified with 3.0% (m/v) agar with final pH 5.8.
Callus induction and plantlet regeneration
After co-cultivation in the dark for 48 h at 21 °C, these hypocotyl sections were transferred into a triangular flask containing CIM. CIM was made of modified MS salts and B5 vitamins supplemented with 0.05 mg/L IAA, 0.05 mg/L KT, 0.05 mg/L 2, 4-D and 25.0 g/L glucose. The pH of the medium was 5.8. The solidified agent (2.0 g/L Gelrite gellan gum) was added to the medium and then CIM was autoclaved at 121 °C for 15 min. To restrain A. tumefaciens, cephalosporin (100 mg/L) was added after autoclaving, and kanamycin (50 mg/L) was added at the same time to select GFP positive calli.
After 4–6 weeks of subculture, explants formed loose and friable calli at 28 °C (Figure 2B). Embryogenic calli were induced and subcultured on SECIM, which contained modified MS salts, B5 vitamins, 0.08 mg/L IAA, 0.16 mg/L KT and 25.0 g/L glucose (pH 6.5). Before autoclaving, Gelrite gellan gum (2.2 g/L) was added to the medium to solidify the medium. Kanamycin (50 mg/L) was added after autoclaving. Calli were transferred onto fresh medium every 4 weeks.
Germination of somatic embryos was carried out on SEGM, which was composed of modified MS salts and B5 vitamins, 0.1 mg/L IAA, 0.2 mg/L 6-BA, and 25.0 g/L sucrose (pH 6.8). Gelrite gellan gum (2.2 g/L) was added before autoclaving (121 °C for 15 min). Germinated embryos (Figure 2H) were transferred to fresh SEGM every 4–6 weeks.
Molecular analyses
Genomic DNA was isolated from untransformed CCRI24 plant and transgenic callus via a modified cetyltrimethylammonium bromide (CTAB) method. PCR analysis for EGFP gene was carried out with primers 5′-GTGAGCTCAGATTTGTAGAGAGAGACTGGT-3′ (forward) and 5′-ATCCCGGGTCACCATTACGAACGATA-3′ (reverse). For further Southern blotting analysis of positive PCR lines, about 50–60 μg genomic DNA were digested with BamH I and Sac I to release a 750 bp fragment containing the coding region of the EGFP gene. A PCR-amplified EGFP cDNA fragment was labeled by digoxigenin and served as the probe to hybridize with DNA samples on the blot (DIG Probe DNA Labeling and Detection Starter Kit I; Roche GmbH, Mannheim, Germany).
Regular light microscopy and fluorescent microscopy
For histological observation, materials were fixed with 4% (v/v) gluteraldehyde in a 0.1 M phosphate buffer (pH 7.2) overnight (or longer), then dehydrated in an ethanol series and embedded in paraffin. About 10 μm thick sections were cut using a Leica microtome (Leica Instruments GmbH, Hubloch, Germany). Sections were stained with safranine fast green and mounted in synthetic resin. Pictures were taken using an Olympus BH-2 microscope.
Fresh tissue was collected from transgenic lines and mounted on microscopic slides just prior to visualization. GFP fluorescence was observed with a fluorescence microscope (Axioskop 40, Carl Zeiss, Jena, Germany) under 488 nm excitation and 510 nm emissions. Photographs were recorded with an Axiocam MRc digital camera (Zeiss) and analyzed by the AxioVision 4 program.
Electron microscopy
Calli were fixed in 4% (v/v) gluteraldehyde in a 0.1 M phosphate buffer (pH 7.2) overnight, rinsed three times in the same buffer, and fixed in 1.0% (w/v) OsO4 (Structure Probe, Inc., West Chester, PA, USA) in phosphate buffer for 3 h. Tissue samples were dehydrated in a series of ethanol gradient solutions. For transmission electron microscopy (TEM), samples were infiltrated and embedded in Spurr's resin (Spurr 1969). Ultra-thin sections (70–90 nm) were cut using a glass knife on an Ultramicrotome Leica Ultracut EM UC6 (Leica) and picked up on a copper grid. Sections were double stained with saturated uranyl acetate and lead citrate, and examined with an H-7500 electron microscope (Hitachi, Tokyo, Japan). For scanning electron microscopy (SEM), samples were critical point dried in liquid CO2 at the HCP-2 critical point drier (Hitachi) and mounted on steel plates, and coated with gold palladium (EIKO-IB5, Eiko Electric Industrial, Ibaragi, Japan). Samples were observed using a Hitachi S-530 Scanning Electron Microscope (Hitachi).
(Handling editor: Qian Qian)
Acknowledgements
We thank Lihua Ma and Tianping Suo of the Cotton Institute, Chinese Academy of Agricultural Sciences, for their technical assistance with SEM, and Xin Den of the Institute of Botany, the Chinese Academy of Sciences for the pGJ280 vector.




