Microsatellite DNA Variation of the Gametophyte Clones Isolated from Introduced Laminaria japonica (Phaeophyta) and L. longissima of China and Varieties Derived from them
Supported by The Key Laboratory of Mariculture of Chinese Ministry of Education, Ocean University of China (200405), and Provincial Department of Science and Technology of Shandong (2005GG3205190).
Abstract
The variation of 90 Laminaria gametophyte clones representing the introduced Laminaria japonica (Group 1) and Laminaria longissima (Group 2), the varieties of L. japonica (Group 3) and the varieties derived from interspecific hybrids (Group 4) was determined with 18 microsatellite markers. The allelic diversity and Nei's gene diversity of Group 1 were significantly higher than those of Group 2 (2.9 vs. 1.8 and 0.414 vs. 0.161, respectively), demonstrating that the variation of the introduced L. japonica is richer than that of L. longissima. Both allelic diversity and Nei's gene diversity of Group 3 were lower than those of Group 1, indicating that only a portion of variation of L. japonica was incorporated into the varieties of L. japonica. Significant genetic differentiation was detected between four groups and between female (Population 1) and male (Population 2) gametophyte clones in each group. The variation among groups accounted for 39.95%, while that among populations accounted for 21.65% of the total. The genetic distance between Group 1 and Group 4 was obviously longer than that between Group 2 and Group 4 (0.686 vs. 0.291), indicating that maternal gametophyte clone contributed more variation to the hybrids than the paternal gametophyte clone did.
Laminaria japonica was introduced into China for cultivation in 1927. With the innovation of summer-sporeling-raising (sporophyte-seedling-raising) technique (Tseng et al. 1955), Laminaria cultivation of China evolved rapidly into the largest mariculture industry in the world (Tseng et al. 1984; Zemke-White et al. 1999; Tseng 2001). Breeding and application of a set of varieties of L. japonica also played crucial roles in the development of the Laminaria mariculture industry of China (Wu and Lin 1987). In recent decades, more and more elite Laminaria varieties and Laminaria hybrids have been bred using gametophyte-cloning and gametophyte-clone-hybridization techniques (Tom 1992; Wang 1994). Danza No.10, a Laminaria hybrid, was bred in 1985. Although not used in cultivation, it marked the initiation of Laminaria hybrid development in China (Fang et al. 1985). After the introduction of L. longissima, variety 901 was bred by crossing female gametophyte clones of L. longissima with male ones of L. japonica followed by self-crossing and desirable trait targeted selection, which held significantly higher yield potential and stress tolerance than the varieties of L. japonica (Zhang et al. 2006). After the hybrid-sporeling-raising technique was established (Li et al. 2003a, 2003b; Li et al. 1999; Zhang et al. 2005), Dongfang No. 2 (a hybrid of a female gametophyte clone of L. longissima and a male gametophyte clone of L. japonica) was developed and cultivated on trial (Li et al. 2006).
At present, Laminaria gametophyte clones are the entities of germplasm resources conserved indoors, and the parental materials of the Laminaria variety and hybrid breeding. China has constructed a few germplasm stocks of Laminaria gametophyte clones. A rich collection of Laminaria gametophyte clones were collected and conserved indoor. Morphological, physiological and molecular markers, for example, random amplified polymorphic DNA (RAPD), amplified fragment length polymorphism (AFLP) and Inter-simple sequence repeat (SSR), have been used to evaluate the variation of conserved Laminaria gametophyte clones (He et al. 2003; Wang et al. 2004, 2005a; Shi et al. 2005). However, diverse and highly variable microsatellite DNA markers have not been used for this purpose. In order to effectively conserve and exploit Laminaria gametophyte clones, 18 polymorphic microsatellite DNA markers were developed and used to determine the diversity of the gametophyte clones isolated from introduced L. japonica and L. longissima of China and varieties derived from them in this study.
Results
Development of Laminaria microsatellite DNA marker
About 30% of colonies of the library contained inserts with microsatellite DNA motifs at a middle position. 167 colonies were sequenced commercially with the automatic sequencer ABI3730 (Sangon, Shanghai, China). Of about 138 sequences obtained 65 most desirables (microsatellite DNA located at a middle position and the flanking regions of both sides were long enough) were used to design the primers as the first trial. Unfortunately, only 18 pairs of primers detected polymorphism in the gametophyte clones isolated from either L. japonica (Group 1) or L. longissima (Group 2) (Tables 1 and 2). In the total sample, the number of alleles per locus ranged from two to seven with an average of 3.9, and the Nei's gene diversity ranged from 0.369 (SH8 and H180) to 0.753 (H120) with an average of 0.554 (Table 3). It was also found that Laminaria microsatellite DNA usually was short and tended to be imperfect (Table 2). Very interestingly, 13 loci were found to be independent of each other in Group 1 in linkage disequilibrium testing, while only one locus was found to be independent in Group 2.
| Source | Female (P1) | Male (P2) |
|---|---|---|
| Laminaria japonica (G1) | 14 | 10 |
| L. longissima (G2) | 14 | 10 |
| Varieties of L. japonica (G3) | 14 | 10 |
| Varieties derived from interspecific hybridsa (G4) | 9 | 9 |
- aThese varieties were bred by hybridizing the female gametophyte clones isolated from L. longissima with the male gametophyte clones isolated from L. japonica and L. longissima, followed by self-crossing of the first filial generation, selection of the most desirable individuals and five to six extra rounds of self-crossing and selection.
| Locus | Accession no. | SSR motif | Primer sequence (5′→3′) | Ta (°C) | Es (bp) |
|---|---|---|---|---|---|
| SH 8 | DQ978335 | (AC)7 | AGAATCGGCACGAACACT a | 53 | 332 |
| CAAACACGAACGACGAAG | |||||
| H 21 | DQ978341 | (AC)10 | AACGCATTGGCTGTCCTTG | 50 | 378 |
| CCACTGCTGGTGACAACTATGA | |||||
| HSSR 18 | DQ978337 | A7(CA)5CG(CA)3 | CCGTCTACCGCTGTATTGTGA | 57 | 364 |
| CGAAAGCATAAGGACGGT | |||||
| H 1 | DQ978338 | (CA)7 | CAACTCAACTACTGCCACCTA | 59 | 341 |
| GTCCTCATCCGTTTCGTC | |||||
| SH 6 | DQ978334 | (TA)5GTAGC(TA)5 | CACCACGCCATACTTCCG | 55 | 322 |
| TCACACCATACTACTGCTCCG | |||||
| H 123 | DQ978346 | (ACAAC)5(ACAAA)6 | AAAGGTCGATAAGCTCGCAGTT | 59 | 379 |
| GCGTTGTTTCGCAAAGTGATT | |||||
| H 10 | DQ978340 | (CAA)9 | TATCCCGTTCGTTCCACTC | 57 | 387 |
| CGACCCTAATAAGCTCTACCT | |||||
| H 105 | DQ978344 | (AC)6 | AGCAACAGAAGCAGCCCAGAG | 57 | 389 |
| GAGCAGACCCAAAGCCAGACA | |||||
| H 120 | DQ978345 | (AC)9 | AAAAGGTGAAGGGATTCGTCG | 59 | 241 |
| CTGAGCAGATTTATGTGGAGCG | |||||
| H 2 | DQ978339 | (AC)11 | CGTGGTAGAACTGCGTAGCG | 51 | 161 |
| AGTGACATCGTTGCCCGTAT | |||||
| H 45 | DQ978342 | (ACAAC)5(ACAAA)6 | TCTTACCCGACTGACCGTGAC | 59 | 290 |
| CTCATCGGTTTTCCCCAAGT | |||||
| H 49 | DQ978343 | (AC)39 | TGGGTATGATGGATGTCGC | 59 | 367 |
| CAATAATAGCATGGCCGTAA | |||||
| H 139 | DQ978348 | (TC)4(AC)7 | CAGCATTGGGCAGGTGAAGGT | 55 | 141 |
| GACGGGACGGCAGTCAAGAAA | |||||
| H 140 | DQ978349 | (CA)6 | ACGACGAAGGGAGAAGAATAA | 52 | 395 |
| TGAGGAGACTGACTGGCAATA | |||||
| H 180 | DQ978352 | (AC)8 | GAGAGTACATGGTGTCCCTAAG | 54 | 233 |
| GCCTCATATGTGTTCGTTTT | |||||
| H 162 | DQ985173 | (AC)8(TA)5 | AGCAAAGAAGGGCGAAGATAA | 60 | 365 |
| GGTTGGAATCATATTTGGCGT | |||||
| H 189 | DQ978354 | (CA)19 | AGAAGAACGGGGCAAAGGC | 59 | 302 |
| CGCCCTTGTTTGGTCGTGT | |||||
| H 144 | DQ978350 | (AC)8 | TCTGCTGATGCTGGTATGGTG | 52 | 257 |
| AAATATCTGCAAAACTAAGGGGC | |||||
- aForward primer (above) and reverse primer (bottom); Es, expected size (the length flanked by primers and including primers); Ta, annealing temperature.
| Locus | G1 | G2 | G3 | G4 | Total | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P1 | P2 | Subtotal | P1 | P2 | Subtotal | P1 | P2 | Subtotal | P1 | P2 | Subtotal | ||
| SH8 | 2 | 2 | 2 | 1 | 1 | 1 | 2 | 2 | 2 | 1 | 1 | 1 | 2 |
| 0.245 | 0.480 | 0.375 | 0.000 | 0.000 | 0.000 | 0.245 | 0.320 | 0.278 | 0.000 | 0.000 | 0.000 | 0.369 | |
| H21 | 1 | 2 | 2 | 1 | 1 | 1 | 3 | 2 | 3 | 1 | 1 | 1 | 3 |
| 0.000 | 0.480 | 0.278 | 0.000 | 0.000 | 0.000 | 0.449 | 0.320 | 0.403 | 0.000 | 0.000 | 0.000 | 0.619 | |
| HSSR18 | 2 | 2 | 2 | 1 | 1 | 1 | 2 | 2 | 2 | 1 | 1 | 1 | 3 |
| 0.245 | 0.320 | 0.278 | 0.000 | 0.000 | 0.000 | 0.490 | 0.480 | 0.486 | 0.000 | 0.000 | 0.000 | 0.631 | |
| H1 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 1 | 2 | 1 | 2 | 2 |
| 0.490 | 0.320 | 0.444 | 0.490 | 0.320 | 0.486 | 0.000 | 0.000 | 0.000 | 0.444 | 0.000 | 0.444 | 0.444 | |
| SH6 | 3 | 2 | 4 | 2 | 2 | 3 | 1 | 2 | 2 | 2 | 1 | 3 | 5 |
| 0.612 | 0.320 | 0.583 | 0.490 | 0.480 | 0.625 | 0.000 | 0.480 | 0.278 | 0.444 | 0.000 | 0.611 | 0.578 | |
| H123 | 4 | 3 | 4 | 2 | 1 | 2 | 2 | 2 | 3 | 2 | 1 | 3 | 5 |
| 0.694 | 0.640 | 0.736 | 0.245 | 0.000 | 0.153 | 0.408 | 0.320 | 0.403 | 0.444 | 0.000 | 0.611 | 0.741 | |
| H10 | 4 | 3 | 5 | 3 | 1 | 3 | 2 | 2 | 2 | 2 | 1 | 2 | 7 |
| 0.612 | 0.560 | 0.611 | 0.449 | 0.000 | 0.292 | 0.490 | 0.480 | 0.486 | 0.444 | 0.000 | 0.278 | 0.626 | |
| H105 | 2 | 1 | 2 | 1 | 1 | 1 | 2 | 2 | 2 | 1 | 1 | 1 | 2 |
| 0.245 | 0.000 | 0.153 | 0.000 | 0.000 | 0.000 | 0.245 | 0.320 | 0.278 | 0.000 | 0.000 | 0.000 | 0.498 | |
| H120 | 3 | 2 | 3 | 2 | 1 | 2 | 1 | 2 | 2 | 3 | 2 | 4 | 7 |
| 0.612 | 0.480 | 0.625 | 0.245 | 0.000 | 0.153 | 0.000 | 0.320 | 0.153 | 0.667 | 0.444 | 0.722 | 0.753 | |
| H2 | 3 | 2 | 4 | 2 | 1 | 2 | 2 | 3 | 4 | 1 | 1 | 1 | 6 |
| 0.449 | 0.480 | 0.514 | 0.245 | 0.000 | 0.153 | 0.408 | 0.560 | 0.694 | 0.000 | 0.000 | 0.000 | 0.680 | |
| H45 | 3 | 2 | 4 | 3 | 1 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 5 |
| 0.449 | 0.320 | 0.417 | 0.449 | 0.000 | 0.292 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.473 | |
| H49 | 3 | 2 | 3 | 1 | 2 | 2 | 1 | 2 | 2 | 2 | 1 | 2 | 4 |
| 0.449 | 0.480 | 0.542 | 0.000 | 0.320 | 0.153 | 0.000 | 0.320 | 0.153 | 0.444 | 0.000 | 0.444 | 0.612 | |
| H139 | 3 | 2 | 4 | 1 | 1 | 1 | 2 | 1 | 3 | 1 | 2 | 3 | 4 |
| 0.571 | 0.480 | 0.597 | 0.000 | 0.000 | 0.000 | 0.245 | 0.000 | 0.569 | 0.000 | 0.444 | 0.611 | 0.516 | |
| H140 | 2 | 2 | 3 | 2 | 1 | 2 | 1 | 2 | 3 | 2 | 1 | 2 | 4 |
| 0.408 | 0.480 | 0.500 | 0.245 | 0.000 | 0.153 | 0.000 | 0.320 | 0.542 | 0.444 | 0.000 | 0.444 | 0.590 | |
| H180 | 1 | 1 | 1 | 2 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 2 |
| 0.000 | 0.000 | 0.000 | 0.245 | 0.000 | 0.153 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.369 | |
| H162 | 1 | 2 | 2 | 1 | 1 | 1 | 3 | 1 | 3 | 2 | 1 | 2 | 3 |
| 0.000 | 0.480 | 0.278 | 0.000 | 0.000 | 0.000 | 0.571 | 0.000 | 0.542 | 0.444 | 0.000 | 0.444 | 0.497 | |
| H189 | 1 | 1 | 1 | 2 | 2 | 2 | 1 | 2 | 2 | 2 | 3 | 3 | 3 |
| 0.000 | 0.000 | 0.000 | 0.245 | 0.320 | 0.278 | 0.000 | 0.320 | 0.153 | 0.444 | 0.667 | 0.611 | 0.484 | |
| H144 | 3 | 3 | 4 | 1 | 1 | 1 | 2 | 2 | 3 | 1 | 2 | 3 | 4 |
| 0.449 | 0.560 | 0.514 | 0.000 | 0.000 | 0.000 | 0.245 | 0.320 | 0.611 | 0.000 | 0.444 | 0.611 | 0.497 | |
| N | 14 | 10 | 24 | 14 | 10 | 24 | 14 | 10 | 24 | 9 | 9 | 18 | 90 |
| Average | 2.4 | 2.0 | 2.9 | 1.7 | 1.2 | 1.8 | 1.7 | 1.8 | 2.3 | 1.6 | 1.3 | 2.0 | 3.9 |
| 0.363 | 0.382 | 0.414 | 0.186 | 0.080 | 0.161 | 0.211 | 0.271 | 0.345 | 0.235 | 0.111 | 0.324 | 0.554 | |
- G1, gametophyte clones isolated from Laminaria japonica; G2, gametophyte clones isolated from Laminaria longissima; G3, gametophyte clones isolated from the varieties of L. japonica; G4, gametophyte clones isolated from the varieties derived from interspecific hybrids; N, number of gametophyte clones used; P1, female gametophyte clones; P2, male gametophyte clones in each group.
Microsatellite DNA variation
As listed in Table 3, the allelic diversity (mean number of alleles) of Group 1 was significantly higher than that of Group 2 (2.9 vs. 1.8, P < 0.05). Similarly, the Nei's gene diversity of Group 1 was also significantly higher than that of Group 2 (0.414 vs. 0.161, P < 0.05). Both allelic diversity and Nei's gene diversity of Group 3 were lower than those of Group 1. Both allelic diversity and Nei's gene diversity of Group 4 were lower than those of Group 1 but higher than those of Group 2. In addition, significant differences of either allelic diversity or Nei's gene diversity or both were observed between female (Population 1) and male (Population 2) gametophyte clones in each group. Significant genetic differentiation was detected between groups and between populations in each group (P < 0.05) (Table 4). The variation among groups accounted for 39.95%, while that among populations accounted for 21.65% of the total (Table 5).
| Gametophyte clones isolated from | G1 | G2 | G3 | Between P1 and P2 each group |
|---|---|---|---|---|
| Laminaria japonica (G1) | 0.155 | |||
| L. longissima (G2) | 0.623 | 0.178 | ||
| Varieties of L. japonica (G3) | 0.283 | 0.657 | 0.439 | |
| Varieties derived from interspecific hybrids (G4) | 0.446 | 0.451 | 0.445 | 0.615 |
- G1, gametophyte clones isolated from Laminaria japonica; G2, gametophyte clones isolated from Laminaria longissima; G3, gametophyte clones isolated from the varieties of L. japonica; G4, gametophyte clones isolated from the varieties derived from interspecific hybrids; P1, female gametophyte clones; P2, male gametophyte clones in each group.
| Source of variation | Percentage | Fixation index | F-statistics | Possibility |
|---|---|---|---|---|
| Among groups | 39.95 | 0.616 | FCT | 0.000 |
| Among populations | 21.65 | 0.360 | FSC | 0.000 |
| Within populations | 38.41 | 0.399 | FST | 0.009 |
The differentiation was also reflected by Nei's genetic distances (Table 6). The distances between Group 1 and Group 2 and between Group 2 and Group 3 reached 1.142 and 1.036, respectively, while that between Group 1 and Group 3 was only 0.278. It was very interesting to note that the genetic distance between Group 1 and Group 4 was obviously longer than that between Group 3 and Group 4 (0.686 vs. 0.291). The genetic distances between groups and between populations in each group were more clearly reflected in Figure 1, where female and male clones in each group were clustered first, then Group 1 and Group 3 clustered, and finally Group 3 and Group 4 clustered.
| Gametophyte clones isolated from | G1 | G2 | G3 | Between P1 and P2 each group |
|---|---|---|---|---|
| Laminaria japonica (G1) | 0.123 | |||
| L. longissima (G2) | 1.142 | 0.037 | ||
| Varieties of L. japonica (G3) | 0.278 | 1.036 | 0.296 | |
| Varieties derived from interspecific hybrids (G4) | 0.686 | 0.291 | 0.527 | 0.440 |
- G1, gametophyte clones isolated from Laminaria japonica; G2, gametophyte clones isolated from Laminaria longissima; G3, gametophyte clones isolated from the varieties of L. japonica; G4, gametophyte clones isolated from the varieties derived from interspecific hybrids; P1, female gametophyte clones; P2, male gametophyte clones in each group.
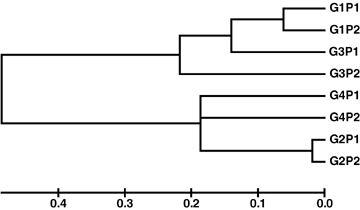
Neighbor-joining tree constructed based on Nei's unbiased genetic distances.G1, gametophyte clones isolated from Laminaria japonica; G2, gametophyte clones isolated from Laminaria longissima; G3, gametophyte clones isolated from the varieties of L. japonica; G4, gametophyte clones isolated from the varieties derived from interspecific hybrids; P1, female gametophyte clones; P2, male gametophyte clones in each group.
The relationship of the four groups was also clearly demonstrated in the assignment test (Table 7). About 33.3% of the gametophyte clones of Group 1 were assigned to Group 3, but not to Group 4. In contrast, 14.3% of the gametophyte clones of Group 2 were assigned to Group 4, but not Group 3. No clones of both Group 3 and Group 4 were assigned back to Group 1 or Group 2. In addition, recent population size reduction (bottleneck effect) was detected in all groups.
| Gametophyte clones isolated from | N | Clones assigned to | |||
|---|---|---|---|---|---|
| G1 | G2 | G3 | G4 | ||
| Laminaria japonica (G1) | 24 | 0.667 | 0.000 | 0.333 | 0.000 |
| L. longissima (G2) | 24 | 0.000 | 0.857 | 0.000 | 0.143 |
| Varieties of L. japonica (G3) | 24 | 0.000 | 0.000 | 1.000 | 0.000 |
| Varieties derived from interspecific hybrids (G4) | 18 | 0.000 | 0.001 | 0.000 | 0.999 |
- G1, gametophyte clones isolated from Laminaria japonica; G2, gametophyte clones isolated from Laminaria longissima; G3, gametophyte clones isolated from the varieties of L. japonica; G4, gametophyte clones isolated from the varieties derived from interspecific hybrids; N, number of gametophyte clones; P1, female gametophyte clones; P2, male gametophyte clones in each group.
Discussion
18 Laminaria microsatellite DNA markers were developed and characterized in the present study and they were the first batch of microsatellite DNA markers of the two Laminaria species. These markers will facilitate the selection of core Laminaria gametophyte clones and their effective conservation and appropriate exploitation. As many as 13 loci were found to deviate from linkage equilibrium in gametophyte clones isolated from L. japonica. This can not be explained completely by physical linkage between markers. Functional linkage may have underlined such phenomenon as was suggested by Flint-Garcia (2003) and Odongo et al. (2006). L. japonica is an introduced species. Some environmental factors, for example, seawater temperature, may have caused concerted selection of some functions and the linkage of genetic markers associated with these functions. Hardy-Weinberg equilibrium (HWE) testing of the markers developed is not applicable; Laminaria gametophytes are haploid and HWE tests the hypotheses of random fusion of male and female gametes and over-dominance or deficiency of heterozygotes.
Laminaria japonica was introduced into China repeatedly during a period of nearly 80 years. Its adaptation to the environment of the Chinese coast may have evolved unique variations. L. longissima was introduced into China later, and was not directly cultured in China. L. japonica should be more diverse than L. longissima although both of them are introduced species. This may explain to some extent why the allelic diversity and the Nei's gene diversity of the gametophyte clones isolated from L. japonica were significantly higher than those of the gametophyte clones isolated from L. longissima. The genetic differentiation of L. japonica and L. longissima was as high as 39.96%. It is expected that elite Laminaria varieties and hybrids can be bred through their hybridization. In fact, the breeding of Laminaria variety 901 (Zhang et al. 2006) and Laminaria hybrid Dongfang No.2 (Li et al. 2006) have proved this inference.
Both allelic diversity and Nei's gene diversity of the gametophyte clones isolated from the varieties of L. japonica were lower than those of the gametophyte clones isolated from L. japonica, indicating that only a portion of variation of L. japonica were incorporated into the varieties of L. japonica. Varieties of L. japonica were bred from L. japonica, which represented only a portion of variation of L. japonica adapted to the culturing condition of the Chinese coast. Some novel variations may have also evolved during cultivation; however, they were not distinguishable from the integrated.
Both allelic diversity and Nei's gene diversity of the gametophyte clones isolated from interspecific hybrids were lower than those of the gametophyte clones isolated from L. japonica, but higher than those of the gametophyte clones isolated from L. longissima. Genetic distance measurement and assignment test showed that the gametophyte clones isolated from interspecific hybrids were genetically closer to those isolated from L. longissima than to those isolated from L. japonica. These observations implied that the female gametophyte clones of L. longissima and the male gametophyte clones of L. japonica contributed unequally to their hybrids and further to the gametophyte clones isolated from these hybrids. In our breeding practice, we have observed that the female and male gametophyte clones contributed differentially to the characteristics of hybrids, and maternal clones contributed more than paternal clones did. In order to achieve an obvious increase in yield, female gametophyte clones of L. longissima were usually used in obtaining hybrids as was done in breeding 901 (Zhang et al. 2006). The mechanism underlining this phenomenon requires further studies.
In the present study, female and male gametophyte clones were found to differentiate significantly. This implied that tow sexes of gametophyte clones may have differentiated to some extent as was suggested by some researchers. Unfortunately, no direct evidence is currently available. In the female gametophytes of Laminariales, a very large X-chromosome has been observed (Evans 1965), which was suggested to mediate sexual differentiation. Sexual differentiation of Laminaria gametophytes is worthy of study in the future; such differentiation directly correlates with breeding. For example, the unequal contributions of female and male gametophyte clones to their hybrids may be explained with such differentiation.
Genetic drift may explain the bottleneck effect detected in all groups; L. japonica and L. longissima are introduced species. The variations they carried are only a portion of the total that exists in nature, and those carried by varieties are a smaller portion of the introduced species.
The gametophyte clones used in the present study represented only introduced L. japonica and L. longissima of China. The comparison of the genetic diversity of these Laminaria and the naturally distributed Laminaria will help us to understand the genetic diversity of the whole Laminaria population. Such comparison will also facilitate the determination of the influence of cultivation on Laminaria variation. If the diversity of the cultured varieties is a small fraction of that available, there may be a large amount of useful diversity in wild populations. Conversely, if the cultured varieties encompass much of the potential diversity, there is no sense in expanding the gametophyte collection to harness this diversity. Although such comparison is informative, however, L. japonica does not naturally habitat the Chinese coast. An international cooperative study is clearly significant.
Materials and Methods
Gametophyte clones
In total, 90 Laminaria gametophyte clones were used in the present study. The number of clones across four groups and two sexes are listed in Table 1. These clones were conserved indoor by Shandong Oriental Ocean Sci-tech Co. Ltd. (Yantai, China), which represented the variation of introduced L. japonica and L. longissima of China and the varieties derived from them. The gametophyte clones were harvested by filtering through a sieve cloth and were stored in 70% ethanol – 15 mM ethylene diamine-tetraacetic acid buffer (EDTA) (pH 8.0) and at −20 °C.
DNA isolation
DNA was isolated from about 100 mg of gametophyte following the protocol described previously (Patwary and Van Der Meer 1994; Wang et al. 2005b; Varela-Alvarez et al. 2006) with minor modifications. In brief, the gametophyte was ground into powder in liquid nitrogen, transferred into 3 mL extraction buffer (2% CTAB, 1.4 mol/L NaCl, 20 mmol/L EDTA, pH 8.0, 100 mmol/L Tris-HCl, pH 8.0 and 2%β-mercaptoethanol), mixed well and incubated at 65 °C for 1 h. The emulsion was mixed with 0.33 volume of 5 M potassium acetate (KAc) (pH 7.5), placed on ice for 30 min and centrifuged at 16 000g for 20 min. The supernatant was extracted with phenol and chloroform (24:1, v:v). The DNA was precipitated with 0.6 volume of isopropanol. The DNA was re-suspended in 300 μL extraction buffer and extracted again with phenol and chloroform after adding 100 μL KAc (pH 7.5). The DNA was precipitated with two volumes of absolute alcohol after adding 0.10 volume of NaAc (pH 5.2), washed with 70% ethanol, air-dried briefly and dissolved in tris(hydroxymethyl)aminomethane ethylene diamine-tetraacetic acid (TE) buffer.
Development of microsatellite DNA markers
A microsatellite DNA containing a short genomic DNA fragment enriched library of L. japonica was constructed following the protocol of Fast Isolation by AFLP of Sequences Containing Repeats (FIASCO) (Vos et al. 1995; Zane et al. 2002). Streptavidin coated on magnetic beads conjugated with the biotin covalently linked to (AC)12 oligonucleotide, which then annealed with single strand microsatellite DNA containing DNA fragments. Microsatellite DNA containing DNA fragments were obtained by separating glass beads from the buffer after hybridization and releasing them from the beads by denaturation. The DNA strand was re-amplified, inserted into a pMD18-T vector (Takara, Dalian, China) and electroporated into Escherichia coli DH5α. The colonies were each subjected to three independent polymerase chain reaction (PCR) screenings in order to check whether microsatellite DNA were located at the middle of the insert. The first PCR reaction was carried out using two universal sequencing primers on the vector. The second reaction was carried out using (AC)12 oligonucleotide and forward sequencing primer, and the third reaction was carried out using (AC)12 oligonucleotide and the reverse sequencing primer. A colony was desirable for sequencing only if the PCR product that was amplified by both sequencing primers was longer than that amplified by either (AC)12 oligonucleotide and the forward sequencing primer or (AC)12 oligonucleotide and the reverse sequencing primer. Those recombinants with desirable inserts were commercially sequenced with the automatic sequencer ABI3730 (Sangon). The sequences with relatively longer microsatellite DNA at the middle and flanking sequences at both sides were used to design primers using Primer Premier 5.0 software (http://www.premierbiosoft.com/primerdesign/).
Genotyping of gametophyte clones
Gametophyte clones were genotyped with the primers designed. PCR reaction was carried out in a 25-μL volume containing one reaction buffer, 2.0 mM MgCl2, 200 μM dNTP (each), 200 μM primers (each direction) and 1 U Taq DNA polymerase. The thermal cycling condition was as follows: pre-denaturation at 94 °C for 4 min, followed by 30 cycles of denaturation at 94 °C for 1 min, annealing at the temperature appropriate for every pair of primers (Table 1) and extension at 72 °C for 1 min and an extra extension at 72 °C for 10 min. PCR product was separated on 6% denaturing acrylamide gel and visualized by silver staining.
Data processing
The number of alleles and Nei's gene diversity of female and male gametophytes (populations) isolated from four different sources of material (groups) and Nei's unbiased genetic distances between them, were calculated using POPGENE version 1.44 (Yeh et al. 2000). Linkage disequilibrium was tested with GENEPOP version 3.4 (Raymond and Rousset 1995) with the significance level of all multiple tests compared with the sequential Bonferroni adjustments (Rice 1989). Significant deviation from linkage disequilibrium was acceptable when the possibility of a multiple test was less than the possibility adjusted with the sequential Bonferroni method. A neighbor-joining tree of eight populations was constructed using MEGA3 (Kumar et al. 2004) based on Nei's unbiased genetic distances. Various F-statistics were calculated using FSTAT 2.9.3 (Goudet 1995) with their significances tested by permutation, of which, FST was the measure of genetic differentiation, which was less biased when the sample size was small (Gaggiotti et al. 1999). The proportion of genetic differentiations among groups and among populations was estimated using hierarchical analysis of molecular variance (AMOVA) implemented in ARLEQUIN 2.000 (Schneider et al. 2000). Recent effective population size reduction (bottleneck effect) was determined using BOTTLENECK software (Cornuet and Luikart 1996). The probability that an individual's genotype would be found in any of the four groups was calculated using the assignment test implemented in STRUCTURE 2.1 (Pritchard et al. 2000).
(Handling editor: Song Ge)




