Behavior of Meiotic Chromosomes in Pinus wallichiana, P. strobus and Their Hybrid and nrDNA Localization in Pollen Mother Cells of the Hybrid by Using FISH
Supported by the National Natural Science Foundation of China (30121003).
Abstract
The complete process of meiosis was investigated in Pinus wallichiana, P. strobus and their artificial hybrid (F1) using microsporocytes. It is revealed that there were slightly lower chiasma frequency, lower ring bivalent frequency, lower meiotic index and distinctly higher frequency of aberrance (chromosomal bridges, fragments or micronuclei) in pollen mother cells (PMCs) of the hybrid (F1) than those of the parental species, which showed a certain degree of differentiation between homologous chromosomes of the two parents. However, relatively higher frequency of ring bivalents and higher meiotic index in all the three entities indicate the great stability of genomes of parental species, and the differentiation of genomes between the two parents must have been slight. Total nineteen signal loci of 18S rDNA were observed in nine bivalents of the hybrid (F1), among which one bivalent bears two loci, while the others have only one. It is suggested that distinct differentiation at genetic level existed in homologous chromosomes of the two parental species, whereas only slight differentiation at karyotypic and genomic levels take place between the parent species.
Pines are very often dominant components of the vegetation throughout temperate parts of the Northern Hemisphere (Richardson 1998). Cytological studies have shown that pines are unusually conservative in karyotypes (Darlington and Wylie 1956; Hizume and Tabaka 1979; Hizume 1988). The karyotypes of all the species investigated in Pinus belong to 1A type, the most symmetrical karyotype in plant species, according to Stebbins' (1971) classification (Sax and Sax 1933; Mehra and Khoshoo 1956; Saylor 1964, 1972, 1983; Muratova 1979, 1980; Chen et al. 1992a, 1992b; Li and Qian 1993; Li 1995). Almost all pine chromosomes are of median centromeres except the shortest pair of chromosomes in some species, where there are slight submetacentric (Sax and Sax 1933), making it difficult to be distinguished merely by analyzing the mitotic features.
Meiotic behavior can be served as the criteria in comparing genomes of different species in the genus Pinus. Ferguson (1904) and Lewis (1908) firstly described the meiotic behavior of pine chromosomes, but found no irregularities. Sax and Sax (1933) reported the average chiasma frequency of 22 species of conifers. Hirayoshi and his coworkers (Hirayoshi et al. 1943) discovered univalents and a complicated quadrivalent in a putative hybrid. Sax (1960) analyzed the meiotic behavior of several hybrids, whose parents came from different areas. These hybrids did not appear significant differentiation from their parents in meiotic behavior, and thereby Sax concluded that structural changes in chromosomes were not important as an isolating mechanism in the speciation of pine. Saylor and Smith (1966) analyzed the meiosis of 21 species and 22 interspecific hybrids in Pinus, and found that there was no difference among the hybrids and their parents in frequency of irregularities.
Since Rayburn and Gill (1985) first introduced the techniques into plant researches, fluorescence in situ hybridization (FISH) has been widely applied in karyotype comparison, chromosome identification, gene localization and interspecific introgression (Baum and Appels 1992; Jiang and Gill 1994; Castilho and Heslop-Harrison 1995; D'Hont et al. 1998; Zoldos et al. 1999). Repeated genes proved to be valuable chromosome markers in FISH localization for a variety of purposes (Sastri et al. 1992; Badaeva et al. 1996; Lubaretz et al. 1996; Brown and Carlson 1997). Among the repeated genes, nrDNA is particularly useful and has been localized in mitotic chromosomes of many genera in the Pinaceae, including Pinus (Gorman et al. 1992; Hizume et al. 1992; Karvonen et al. 1993; Doudrick et al. 1995; Hizume et al. 2002; Liu et al. 2003), Picea (Lubaretz et al. 1996; Brown and Carlson 1997; Hizume et al. 1999; Liu et al. 2003), Larix (Lubaretz et al. 1996), and Pseudotsuga (Hizume and Akiyama 1992; Amarasinghe and Carlson 1998). These studies showed that the number of 18S rDNA loci varied from three to ten pairs, and 5S rDNA loci from two to six pairs among the species of the Pinaceae. Using the multicolor FISH method, including nrDNA markers, Hizume and his coworkers (1999) have clearly identified every pair of the coniferous chromosomes in one slide. Based on locus number and relative location of the nrDNA signals, the information including the relationship between the species could be revealed. However, in Pinus the FISH results reported so far have been all on mitotic chromosomes instead of meiotic ones.
Pinus wallichiana Jacks (Himalayan white pine) is distributed from Southwestern China to South Asia (Fu et al. 1999), while P. strobus L. (Eastern white pine) spreads widely in Northeastern America. The karyotypic formulae of both P. wallichiana (Mehra and Khoshoo 1956; Santamour 1960; Saylor 1983; Ohri and Khoshoo 1986; Mehra 1988) and P. strobus (Ferguson 1901, 1904; Sax and Sax 1933; Santamour 1960; Saylor 1961; Baranec 1979; Love and Love 1980; Saylor 1983) are K(2n) = 24 = 22m + 2sm, in which the shortest pair of chromosomes is of submedian centromere. The artifical hybrid (F1) investigated in our study was created in 1970s by Zhou and Dong.
The present study is focused on observation and analyses of complete process of meiosis of P. strobus, P. wallichiana and their F1 hybrid, and localization of 18S rDNA and 5S rDNA onto the meiotic chromosomes, and further to reveal certain information of relationships among the three entities.
Results
Meiotic chromosome behavior
The complete meiotic processes of P. strobus, P. wallichiana and their hybrid (F1) were observed. The images of meiotic chromosomes are shown in Figure 1.
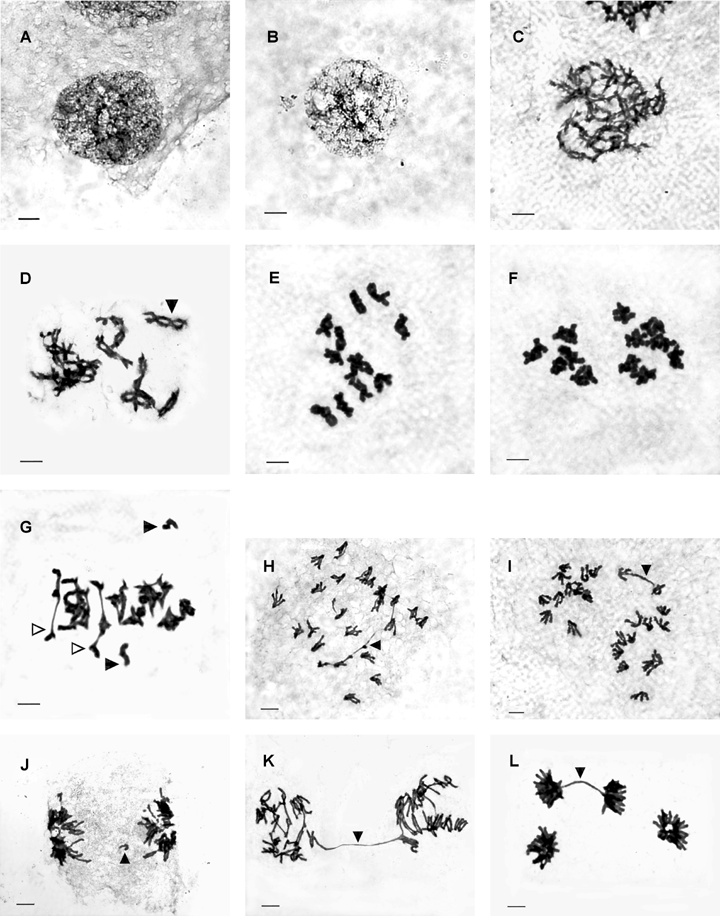
Meotic chromosome bahavior.(A). Leptotene, Pinus strobus.(B). Zygotene, F1.(C). Pachytene, F1.(D). Diplotene, P. wallichiana (arrowhead: chiasma).(E). Diakinesis, P. wallichiana.(F). Diakinesis, P. strobus.(G). Mephase I, F1 (solid arrowheads: univalents; hollow arrowheads: rod bivalents).(H). Anaphase I, P. strobus (arrowhead: inversion bridge).(I). Anaphase I, P. wallichiana (arrowhead: bridge arising from the failure of chiasma terminization).(J). Telophase I, F1 (arrowhead: fragment).(K). Anaphase II, F1 (arrowhead: bridge arising from the first division).(L). Telophase II, P. wallichiana (arrowhead: bridge arising from the second division). Scale bar = 10 μm in all figures.
Prophase I
Prophase I are composed of five sub-stages: leptotene (Figure 1A), zygotene (Figure 1B), pachytene (Figure 1C), diplotene (Figure 1D) and diakinesis (Figure 1E, F).
The chiasmata at diplotene were clearly visible (Figure 1D). The number of chiasmata decreased continuously following the development of chiasma terminization. To ensure the comparability, the number of chiasmata was counted at early diakinesis. The average frequency of chiasma of F1 was 2.24 (Table 1), slightly lower than that of P. wallichiana (2.36) and P. strobus (2.48), confirming those of other conifers reported (Sax 1932, 1933, 1960; Sax and Sax 1933).
| Taxon | Meiotic configuration* | No. of cells | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 12II0 | 11II0 1bII1 | 10II0 2 II1 | 91II0 3 II1 | 8II0 4 II1 | 11II0 2I | 10II0 4I | 10II0 1II1 2I | 9II0 1II1 4I | 9II0 2II1 2I | 8II0 2II1 4I | 8II0 3II1 2I | 7II0 4II1 2I | ||
| Pinus strobus | 68.31 | 20.63 | 6.41 | 1.49 | 0.65 | 0.58 | 0.93 | 0.28 | 0.65 | 0.09 | 1 076 | |||
| P. wallichiana | 88.44 | 10.43 | 0.78 | 0.17 | 0.17 | 2 302 | ||||||||
| F1 | 50.06 | 25.68 | 11.52 | 2.64 | 0.32 | 5.34 | 0.06 | 2.32 | 1.22 | 0.12 | 0.58 | 0.13 | 1 554 | |
- *II0, ring bivalent; II1, rod bivalent; I, univalent.
Metaphase I
Three pairing patterns of homologous chromosomes were observed in PMCs (Figure 1E–G): (1) ring bivalents (II0); (2) rod bivalents (II1); (3) univalents. Meiotic configurations (Table 1) were firstly formed according to the distribution of ring and rod bivalents and univalents in PMCs. Average configuration and paring index were calculated in Table 2.
| Taxon | Average configurations* | Meiotic index (%) | ||
|---|---|---|---|---|
| IIo | II1 | I | ||
| Pinus strobus | 11.750 0 | 0.219 4 | 0.030 6 | 97.62 |
| P. wallichiana | 11.935 7 | 0.062 6 | 0.001 7 | 99.46 |
| F1 | 11.574 3 | 0.325 9 | 0.099 7 | 96.45 |
- *II0, ring bivalent; II1, rod bivalent; I, univalent.
Twelve meiotic configurations were observed in PMCs of the F1, more than those in the two parents (Table 1). The dominating configurations in F1 were 12II0, 11II0+1II1 and 10II0+2II1, composing of 87.26% PMCs in total. Ten meiotic configurations were observed in P. strobus, in which the dominating conifigurations were also 12II0, 11II0+1II1 and 10II0+2II1, composing of 95.35% PMCs in total. Only five meiotic configurations were observed in P. wallichiana, in which the dominating configurations were 12II0 and 11II0+1II1, composing of 98.87% PMCs in total.
The frequency of ring bivalents and meiotic index of F1 (11.57 and 96.45%) were lower than those of the two parents (Table 2), which indicates a certain degree of differentiation of homologous chromosomes between the parental genomes. However, in all the three entities, the frequency of ring bivalents exceeded 11.57, and the meiotic index exceeded 96.45%, which indicates that the differentiation of homologous chromosomes between the parental genomes was slight.
Anaphase I and telophase I
Chromosome bridges, fragments or micronuclei were observed at anaphase I in all the three entities (Figure 1H–J). The chromosome bridges arose primarily from two processes (Saylor and Smith 1966): (1) nondisjunction of bivalents due to failing of chiasma terminization (Figure 1H); (2) crossing-over occurred in inversion regions (Figure 1I). In the former, the distal portions of the chromosome arms were clearly visible, but in the inversion bridges, the chromatin strands extending across the cell were uniform. Both at anaphase I and at telophase I, the frequency of aberrance (including chromosome bridges, fragments and micronuclei) in F1 (26.67% and 6.05%) was obviously higher than those in P. wallichiana (5.24% and 0) and P. strobus (11.3% and 1.63%), which also shows a certain differentiation of homologous chromosomes between the two parental genomes (Table 3).
| Taxon | Frequency in the first division | Frequency in the second division | |||||
|---|---|---|---|---|---|---|---|
| Anaphase I | Talephase I | Average | No. of cells | % | |||
| No. of cells | % | No. of cells | % | ||||
| Pinus strobus | 191 | 5.24 | 246 | 0 | 2.62 | 1 935 | 0.103 4 |
| P. wallichiana | 115 | 11.30 | 306 | 1.63 | 6.47 | 658 | 0.455 9 |
| F1 | 315 | 26.67 | 471 | 6.05 | 16.36 | 870 | 1.149 4 |
- *Meiotic aberrance including bridges, fragments or micronuclei.
Second division
Chromosome bridges in the second division arose from two ways: (1) formed at the first division and remained through the second division (Figure 1K); (2) formed at anaphase II when the chromatids failed in separation (Figure 1L). The frequency of aberrance in the second division of P. strobus, P. wallichiana and F1 was 0.46%, 0.10% and 1.14% respectively, markedly lower than those in the first division (Table 3).
nrDNA's FISH localization
5S rDNA
A pair of 5S rDNA signalized loci was observed in each of two bivalents (Figure 2B). There was no difference of 5S rDNA signalized loci between the two parental chromosomes. A pair of 5S rDNA signalized loci was also observed co-existing with 18S rDNA signals on the same bivalent near the centromere (Figure 2B, 2D).
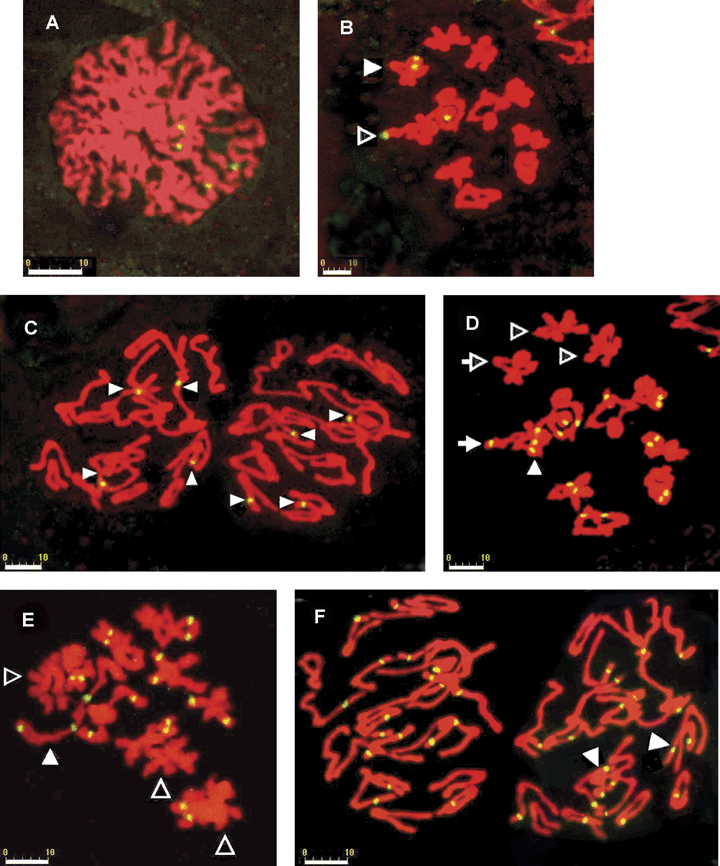
FISH localization of nrDNA in PMCs of F1.(A)–(C) showing 5S rDNA signalized loci.(A). Zygotene, showing 4 sites of 5S rDNA signals. (B). Diakinesis, solid arrowhead: a bivalent with a pair of 5S rDNA signalized loci; hollow arrowhead: a bivalent with a pair of 5S rDNA signalized loci co-existing with a pair of 18S rDNA signalized loci (shown in D). (C). Anaphase II, arrowheads: 5S rDNA signalized loci.(D)–(F) showing 18S rDNA signalized loci.(D). Diakinesis, solid arrowhead: a bivalent with three 18S rDNA signalized loci; solid arrowhead with a tail: a bivalent with a pair of 18S rDNA signalized loci co-existing with a pair of 5S rDNA signalized loci (shown in B); hollow arrowhead with a tail: a bivalent without 18S rDNA signalized loci, but with a pair of 5S rDNA signalized loci (shown in B); hollow arrowheads without tail: the bivalent with neither 18S rDNA signalized loci nor 5S rDNA signalized loci; all the bivalents without any arrowheads have a pair of 18S rDNA signalized loci.(E) Diakinesis, solid arrowhead: a bivalent with three 18S rDNA signalized loci; hollow arrowheads: the bivalents without 18S rDNA signalized loci; all the bivalents without any arrowheads have a pair of 18S rDNA signalized loci.(F) Anaphase II, arrowheads: chromosomes with two 18S rDNA signalized loci.Scale bar = 10 μm in all figures.
18S rDNA
In PMCs of F1, 19 signalized loci of 18S rDNA in total were detected in nine bivalents (Figure 2D–F), in which eight bivalents bear a pair of signalized loci, and a bivalent had three signalized loci with one locus in one chromosome and two in the other. Differentiation in 18S rDNA signalized loci between the homologous chromosomes in F1 indicates the heterozygosity of the two parental chromosomes. In spite of the evident differentiation of 18S rDNA signalized loci, the homologous chromosomes can get tied tightly with each other to form ring or rod bivalents. The differentiation of homologous chromosomes should exist in more minute structures such as in DNA sequences.
Discussion
Meiotic behavior in hybrid in Pinus
As pointed out earlier, karyotypes of pines are unusually conservative (Darlington and Wylie 1956; Hizume and Tanaka 1979; Hizume 1988; Li 1995), with little differentiation among species. It is difficult to distinguish pine chromosomes when merely relied on mitotic morphological characters. Analyses of meiotic behavior will provide constitutional information between homologous chromosomes or genomes. The previous reports on meiosis revealed unusual conservative feature of Pinus chromosomes (Furguson 1904; Lewis 1908; Sax and Sax 1933; Hirayoshi et al. 1943; Sax 1960; Saylor and Smith 1966). There was relatively lower frequency of irregularities in both hybrids and their parents, and there was no obvious difference between species and their hybrids in frequency of either chiasma numbers or meiotic aberrances (Sax and Sax 1933; Sax 1960; Salylor and Smith 1966).
As shown in this study, relatively higher frequency of ring bivalents and meiotic index in all the three entities indicate that the differentiation of homologous chromosomes between the two parents is slight, although the two parental species have been separated for long time, geographically disjuncted distribution in different continents since the end of the Mesozoic (Price et al. 1998; Richardson 1998). However, a lower frequency of ring bivalents and meiotic index were observed in F1 than those in the parents, which indicates that a certain degree of differentiation of homologous chromosomes exists between the two parents.
rDNA localization on meiotic chromosomes by using FISH
Some methods were used for the chromosome identification in Pinus, such as the examination of the secondary constriction (Pederick 1970), C-banding (Borzan and Papes 1978), G-banding (Drewry 1982), and fluorescence banding (Hizume et al. 1983, 1989, 1990; Hizume 1987; Hizume and Akiyama 1992). These procedures are not favorable in many species because of their low reproducibility. Recently, fluorescence in situ hybridization has been used in the localization of rDNA (Gorman et al. 1992; Hizume et al. 1992; Karvonen et al. 1993; Doudrick et al. 1995; Lubaretz et al. 1996; Jacobs et al. 2000; Liu et al. 2003) and other repetitive DNA sequences (Fuchs et al. 1995; Hizume et al. 2000, 2001) in Pinus. However, all these studies above were based on the mitotic chromosomes. In PMCs, the difference of FISH signal location in homologs can be observed conveniently in bivalents. In this study, we localized nrDNA in the meiotic chromosomes of gymnosperm using FISH for the first time. In our investigation, the diversity of 18S rDNA signalized location was found in homologous chromosomes of an interspecific hybrid, showing that the homologous chromosomes derived from different parental species were obviously diversified.
Materials and Methods
Microspores collection and chromosome slide preparation
Microspores of the three entities, P. strobus, P. wallichiana and their F1 hybrid, were collected from trees cultivated in the Beijing Botanical Garden, the Chinese Academy of Sciences (CAS), during April and May in 2002. The materials were firstly fixed in Carnol's Fixation (alcohol: glacial acetic acid = 3:1, v/v) for 24 hours, and then transferred to 75% alcohol, and stored at −20°C. The meiotic preparations were made according to the protocol of Chen and his coworkers (Chen et al. 1979). The microspores were immersed in 1 mol/L HCl for 1 hour, soaked in double distilled water for 30 min and 45% glacial acetic acid for 10 min, and then squashed on slides. The chromosomes were stained in 1:20 Giemsa staining solution (pH = 7.0). Chromosome slides were observed under an optical microscope (Olympus, BH-2). The microphotographs were taken under a Leica microscope (DMRBA) and adjusted using Photoshop 7.0.1 to enhance the contrast. The statistical data were analyzed using EXCELL 2000 and SPSS 10.
Probe preparation
Genomic DNA of P. wallichiana was extracted from silica gel dried needles using the CTAB method following the protocol of Rogers and Bendich (1988) and used as a template for polymerase chain reaction (PCR) amplification. The PCR amplification was conducted in Peltier Thermal Cycler (Biometra), with a reaction volume of 20 μL, containing 20 ng DNA template, 0.2 mmol/L of each dNTP, 2 mmol/L MgCl2, 0.5 U Taq DNA polymerase, and 0.2 μmol/L of each primer described below. 18S rDNA was amplified using following primers: 5′-CTAGAGCTAATACGTGCAAC-3′ and 5′-GATAAGGTTCAGTGGACTTC-3′ (Troitsky et al. 1991). PCR cycles were as follows: 2 min at 95°C for initial denaturation, followed by 34 cycles: 30 s at 94°C, 30 s at 55°C and 2 min at 72°C; The last cycle at 72°C was extended to 8 min. The primers for 5S rDNA amplification were 5′-CGGTGCATTAATGCTGGTAT-3′ and 5′-CCCATCCGTGTACTACTCTC-3′ (Amarasinghe and Carlson 1998). The PCR program for 5S rDNA amplification was almost the same as for 18S rDNA, except the annealing temperature was 60°C with the remaining time of 1 min. The PCR products were purified using a GFX™ PCR DNA and Gel Band Purification Kit (Amersham Biosciences). The purified rDNA preparations were labeled with DIG (digoxigenin-dUTP, Roche) by the random-primed DNA synthesis.
In situ hybridization
The procedure for in situ hybridization followed that of Liu et al. (2003) with slight modification. The chromosome slides were treated with RNase (100 μg/mL, DNase free) at 37°C for 1 h, soaked twice in 2× SSC, each for 5 min, digested with proteinase K (1 μg/μL) at 37°C for 30 min, fixed in 4% paraformaldehyde for 10 min, dehydrated in a series of 70%, 95% and 100% ethanol, and then air-dried. A 20 μL hybridization solution (containing 10% dextran sulfate, 0.1% SDS, 50% formamide, 10 ng/μL sheared salmon sperm DNA, about 2 ng/μL pre-denatured probe and 2× SSC) was added to each slide. The slides were placed in a moisture chamber, denatured at 90°C for 15 min, then immediately put into a hybridization oven, at 37°C overnight. The slides were then washed with 20% formamide (in 0.1× SSC) at 42°C for 10 min, 2× SSC at 37°C for 2× 5 min, 2× SSC at room temperature for 2× 5 min. Before detection, the slides were soaked in washing buffer (0.2% Tween 20 in 4× SSC) at room temperature for 5 min, and then in detecting buffer (5% BSA in washing buffer) at 37°C for 30 min. The DIG-labeled signals were detected using anti-DIG-fluorescence. The chromosomes were counter-stained with 2.5 μg/mL PI for 15 min. The images of signals were observed and scanned under Confoc (BIO-RAD, 1024). The scanned images were adjusted using Photoshop 7.0.1 to enhance the contrast.
(Handling editor: Jin-Zhong Cui)
Acknowledgements
The authors thank Dr. Zhan-Lin LIU and Dr. Qing CAI for valuable instructions in the experiment, and Madam Yu-Mei LIN for the material collection.




