New 16Sr subgroups and distinct single nucleotide polymorphism lineages among grapevine Bois noir phytoplasma populations
Abstract
Bois noir (BN) is an insect-transmitted grapevine yellows disease caused by phytoplasmas belonging to the stolbur subgroup 16SrXII-A. In Italy, increasing prevalence of stolbur phytoplasma strains in vineyards suggests progressive spread of the disease and potential for heavy impacts on the wine industry. In this study, we investigated the genetic diversity of stolbur phytoplasma strains in BN phytoplasma populations. Nucleotide sequences of 16S rRNA genes from stolbur phytoplasma strains affecting vineyards in the Lombardy region of Italy and stolbur phytoplasma 16S rDNA sequences retrieved from GenBank were subjected to virtual restriction fragment length polymorphism analysis. Calculation of virtual restriction similarity coefficients revealed the presence of new subgroups in group 16SrXII (stolbur phytoplasma group). Representative strains of confirmed new subgroups 16SrXII-F (XII-F) and XII-G and tentative new subgroups XII-A1 through XII-A19, XII-H, XII-I, and XII-J as well as known subgroup XII-A were from grapevines; strains representing three additional tentative new subgroups (XII-K, XII-L and XII-M) were from other plant hosts. Nucleotide sequence alignments identified no less than nine genetically distinct 16S rDNA single nucleotide polymorphism lineages from grapevine, indicating a high degree of genetic heterogeneity within BN phytoplasma populations. The findings open new opportunities for in-depth studies of the distribution of grapevine-associated 16SrXII phytoplasma strains in weeds, insect vector populations and grapevines from vineyards located in different geographic areas.
Introduction
Bois noir (BN), known as Legno nero (LN) in Italy and Vergilbungskrankheit (VK) in Germany, is a grapevine yellows (GY) disease that is caused by phytoplasmas in Europe, from Spain to Ukraine (Battle et al., 2000; Milkus et al., 2005), where it induces severe crop losses in almost all varieties used for wine production (Boudon-Padieu, 2003). Results from a recent survey underscored the prevalence of stolbur phytoplasmas in vineyards in Italy (Botti & Bertaccini, 2007), suggesting potential for heavy impacts on Italian viticulture. BN produces typical GY symptoms, including desiccation of inflorescences, reduction of growth, berry shrivel and irregular maturation of wood.
Phytoplasmas are cell-wall-less obligate intracellular parasites belonging to the class Mollicutes. To date, no phytoplasma has been cultured in a cell-free medium; thus, differentiation and classification of phytoplasmas is based on nucleic-acid-based assay techniques (Lee & Davis, 1988; Lee et al., 1992). On the basis of 16S rDNA sequence analysis, the presumptive aetiological agent of BN was identified as ‘Candidatus Phytoplasma solani’, a phytoplasma species belonging to the stolbur subgroup (16SrXII-A) (IRPCM Phytoplasma/Spiroplasma Working Team – Phytoplasma Taxonomy Group, 2004; according to rule 28b of the Bacteriological Code, ‘Candidatus Phytoplasma solani’ is an incidental citation and does not constitute prior citation). Stolbur phytoplasma is transmitted by polyphagous planthoppers of the family Cixiidae (Weintraub & Beanland, 2006) and affects a wide range of wild and cultivated plants. In vineyards, BN phytoplasma is transmitted from plant-to-plant by Hyalesthes obsoletus Signoret (Maixner, 1994; Sforza et al., 1998; Alma et al., 2002), but in wine-growing areas where H. obsoletus is absent, the presence of stolbur phytoplasma implies the existence of alternative vectors (Boudon-Padieu, 2000; Gatineau et al., 2003; Garau et al., 2004; Palermo et al., 2004).
The biological complexity of BN disease has stimulated research on molecular markers of grapevine-affecting stolbur phytoplasma genetic diversity. Seruga Musićet al. (2008) reported genetic diversity in grapevine-associated 16SrXII-A subgroup strains on the basis of single-strand conformation polymorphism (SSCP) analysis of 16S rRNA, tuf (elongation factor Tu), and dnaB genes. Pacifico et al. (2007) found that restriction analysis of the stol-1H10 gene, encoding a putative membrane protein, was a useful marker to type BN phytoplasmas in plants and insects. Analysis revealed that three tuf sequence variants (VK-I, VK-II and VK-III) of ‘Ca. Phytoplasma solani’ were present in BN-diseased grapevines as well as in certain weeds (Langer & Maixner, 2004). Intriguingly, VK-I and VK-II BN phytoplasma strains are differentially distributed in distinct geographic regions in Italy (Pasquini et al., 2007; Quaglino et al., 2007; Riolo et al., 2007).
In the present study, a high degree of genetic heterogeneity among 79 BN phytoplasma strains, detected in north-Italian vineyards from 2004 to 2007, was evidenced by the presence of two new 16SrXII subgroups (16SrXII-F and 16SrXII-G) and of no less than nine genetically distinct 16S rDNA single nucleotide polymorphism (SNP) lineages. The findings expand the classification of group 16SrXII and open new avenues for researching the complex ecology and epidemiology of BN disease.
Materials and methods
Sample collection
Field surveys on the incidence of GY disease were carried out from 2004 to 2007 in 21 vineyards located in Lombardy, northern Italy. Leaf samples were collected from 79 BN-diseased, symptomatic grapevine plants.
Bois noir phytoplasma identification and characterization
Total DNA was extracted from 100 mg of leaf veins per plant using the ‘DNeasy PLANT MINI KIT’ (Qiagen, Hilden, Germany) according to manufacturer’s instructions. Detection of ‘Ca. Phytoplasma solani’ was carried out by means of amplification of 16S rDNA in nested polymerase chain reactions (PCRs) primed by primer pair P1/P7 (Deng & Hiruki, 1991) followed by 16SrI group-specific primer pair R16F1/R16R1 (Lee et al., 1994), and subsequent MseI-restriction fragment length polymorphism (RFLP) assay of amplicons as previously described (Lee et al., 1998). PCRs were performed by using Taq polymerase (Applied Biosystems, Foster City, CA, USA) in an automated thermal cycler (Bio-Rad, Hercules, CA, USA). Presence of PCR amplicons was verified by electrophoresis through 1% agarose gels; restriction fragments were separated by electrophoresis through 5% polyacrylamide gels. DNAs extracted from phytoplasma strains EY1 (‘Ca. Phytoplasma ulmi’, subgroup 16SrV-A), STOL (stolbur group, subgroup 16SrXII-A), and AY1 (‘Ca. Phytoplasma asteris’, subgroup 16SrI-B) served as reference strains. These phytoplasmas were maintained in Madagascar periwinkle (Catharanthus roseus (L.) G. Don). Reaction mixtures containing DNA from healthy Madagascar periwinkle plants and reaction mixtures without DNA template were used as negative controls.
Cloning and sequencing of phytoplasmal 16S rRNA gene
Amplicons from nested PCRs, primed by phytoplasma-universal primer pairs P1/P7 and R16F2n/R16R2 (Gundersen & Lee, 1996) from 79 BN phytoplasma strains were cloned in plasmid vector pCRIITOPO (Invitrogen, Carlsbad, CA, USA) and propagated in Escherichia coli as described (Shuman, 1994). Both strands of cloned inserts were sequenced to achieve at least 4× coverage per base position. DNA sequencing was performed in an ABI PRISM 377 automated DNA sequencer (Applied Biosystems). The nucleotide sequence data were assembled by employing the Contig Assembling program of the sequence analysis software BIOEDIT, version 7.0.0 (http://www.mbio.ncsu.edu/Bioedit/bioedit.html) and deposited in the GenBank database.
Virtual restriction fragment length polymorphism analysis and calculation of similarity coefficients
A total of 99 16S rRNA gene sequences (20 from GenBank and 79 obtained in this work) were trimmed to an approximately 1.25-kb fragment (delimited by R16F2n and R16R2 primer annealing positions, the F2nR2 fragment), as previously described (Wei et al., 2007), and exported to the program pDRAW32 (AcaClone software, http://www.acaclone.com). Each DNA sequence was analysed through an automated in silico restriction assay, and digestion results were plotted on virtual gels as described by Wei et al. (2007). Briefly, each DNA fragment was digested in silico with 17 restriction enzymes used previously in actual enzymatic digestions by Lee et al. (1998): AluI, BamHI, BfaI, BstUI (ThaI), DraI, EcoRI, HaeIII, HhaI, HinfI, HpaI, HpaII, KpnI, Sau3AI (MboI), MseI, RsaI, SspI and TaqI. After in silico restriction digestion, a virtual 3.0% agarose gel electrophoresis image was plotted and captured as a device-independent PDF file. The virtual RFLP patterns were compared and a similarity coefficient (F) was calculated for each pair of phytoplasma strains according to the formula described previously (Nei & Li, 1979; Lee et al., 1998), F = 2Nxy/(Nx + Ny), in which x and y are two given strains under study; Nx and Ny are the total number of bands resulting from digestions by 17 enzymes in strains x and y, respectively; and Nxy is the number of bands shared by the two strains.
Identification and validation of single nucleotide polymorphisms
Nucleotide sequences were compiled in FASTA format, aligned using ClustalX (V1.83) and searched for SNPs. Presence of SNPs in recognition sites for restriction endonucleases was determined by automated in silico restriction digest assays selecting the parameter ‘all restriction enzymes’ in the program pDRAW32, and a virtual electrophoresis gel (3% agarose) pattern of restriction digest products was captured according to Wei et al. (2007). SNPs detected by in silico analyses were validated by actual enzymatic RFLP analyses of representative amplicons; restriction patterns were visualised using 4% agarose gel electrophoresis.
Phylogenetic analysis
16S rRNA gene sequences (Tables 2) were retrieved from the National Center of Biotechnology Information (NCBI) website (http://www.ncbi.nim.nih.gov/BLAST/) and used to construct phylogenetic trees. Minimum evolution analysis was carried out using the neighbour-joining method and bootstrap replicated 1000 times with the software MEGA4 (http://www.megasoftware.net/index.html) (Kumar et al., 2004). Acholeplasma palmae was used as the outgroup.
 |
Results and discussion
Bois noir phytoplasma in grapevines in Lombardy
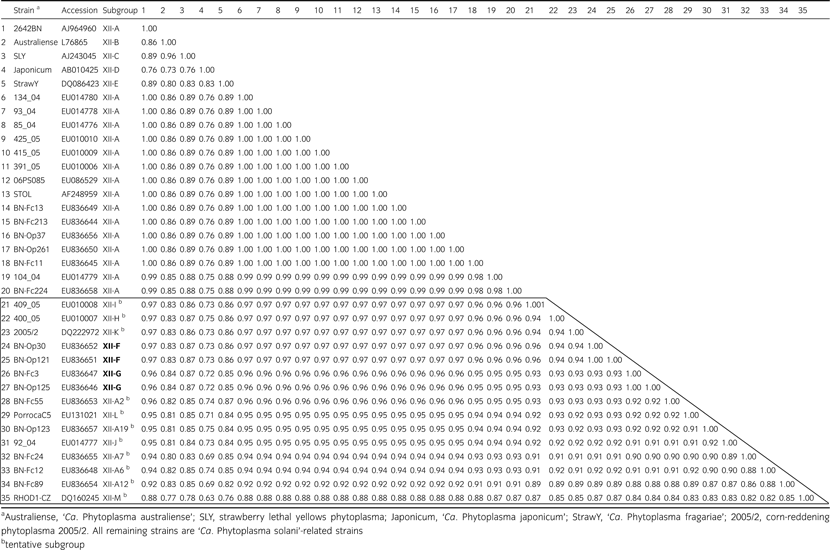
Primer pair R16F1/R16R1, which is known to prime amplification of 16S rDNA from phytoplasmas classified in groups 16SrI and 16SrXII (Lee et al., 1994), primed amplification of DNA from templates derived from all samples studied (data not shown). All amplicons yielded MseI-RFLP patterns that were indistinguishable from one another and from the MseI pattern characteristic of the reference strain STOL, indicating that the strains detected in diseased grapevines were members of group (16SrXII) (Fig. 1). Control PCRs containing template DNA from healthy periwinkle, or water in place of DNA, yielded no observable DNA amplification.
MseI-restriction fragment length polymorphism patterns of polymerase chain reaction amplicons primed by 16SrI group-specific primers R16F1/R16R1. Bois noir (BN)-Fc213, BN-Franciacorta strain 213; BN-Op123, BN-Oltrepo’ pavese strain 123; BN-Fc3, BN-Franciacorta strain 3; BN-Op30, BN-Oltrepo’ pavese strain 30; AY, Aster Yellows phytoplasma; STOL, stolbur phytoplasma; M, molecular marker Φx174 digested by HaeIII (Invitrogen).
New subgroups in group 16SrXII in Italy
Computer-simulated restriction analyses were carried out on F2nR2 fragments from 79 phytoplasma strains involved in BN disease in Lombardy. Visualisation and comparison of virtual gel plotted images revealed 22 different RFLP patterns (data not shown), indicating an unexpectedly high genetic diversity among BN phytoplasma strains in Italy (Table 1). One pattern was exhibited by DNAs from 56 BN phytoplasma strains; this pattern was indistinguishable from that characteristic of strains classified in the ‘Ca. Phytoplasma solani’ subgroup (16SrXII-A) (Fig. 2).
| Area or province | Vineyard | Host | Number of BN strains | 16SrXII subgroupa |
|---|---|---|---|---|
| Franciacorta | Erbusco, Tajardino | Chardonnay | 9 | XII-A (6), XII-A1b (1), XII-A2b (1), XII-G (1) |
| Erbusco, Ronco-Pozzi | Chardonnay | 3 | XII-A (3) | |
| Erbusco, Rampaneto | Chardonnay | 1 | XII-A (1) | |
| Erbusco, podere Pio IX | Chardonnay | 1 | XII-A3b (1) | |
| Rovato, Seriole | Chardonnay | 11 | XII-A (8), XII-A4b (1), XII-A5b (1), XII-A6b (1) | |
| Calino di Cazzago | Chardonnay | 7 | XII-A (5), XII-A7b(1), XII-A8b (1) | |
| Cazzago San Martino | Chardonnay | 4 | XII-A (3), XII-A9b (1) | |
| Fantecolo | Chardonnay | 5 | XII-A (4), XII-A10b (1) | |
| Passirano | Chardonnay | 6 | XII-A (2), XII-A11b (1), XII-A12b (1), XII-A13b (1), XII-A14b (1) | |
| Gussago | Chardonnay | 3 | XII-A (3) | |
| Adro | Chardonnay | 2 | XII-A (2) | |
| Oltrepo’ pavese | Godiasco, Montalfeo | Croatina | 4 | XII-A (2), XII-A15b (1), XII-F (1) |
| Godiasco, Cabanon | Chardonnay | 1 | XII-A16b (1) | |
| Borgo Priolo, Codalunga | Barbera | 4 | XII-A (3), XII-A17b (1) | |
| Chardonnay | 1 | XII-A (1) | ||
| Borgo Priolo, Olesi | Barbera | 1 | XII-A (1) | |
| Cigognola | Cabernet Sauvignon | 1 | XII-A (1) | |
| Montù Beccaria | Barbera | 1 | XII-A (1) | |
| Canevino | Chardonnay | 7 | XII-A (3), XII-A18b (1), XII-A19b (1), XII-F (1), XII-G (1) | |
| Spessa Po | Pinot nero | 2 | XII-A (2) | |
| Pinot grigio | 1 | XII-A (1) | ||
| Pinot nero | 1 | XII-A (1) | ||
| Valtenesi | Bedizzole | Sangiovese | 2 | XII-A (2) |
| Milan | San Colombano al Lambro | Chardonnay | 1 | XII-A (1) |
- a New confirmed subgroups are indicated in bold.
- b New tentative subgroup; numbers of BN strains in each 16SrXII subgroup are given within parentheses.
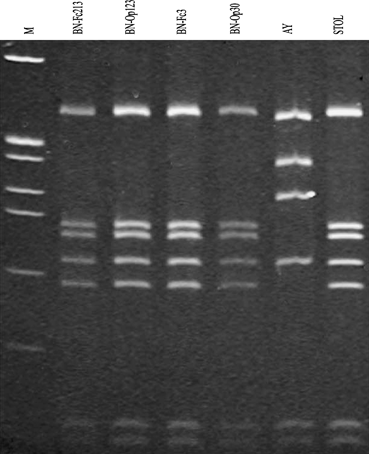
Virtual F2nR2 restriction fragment length polymorphism patterns of representative second ‘australiense’ strains of 16SrXII subgroups. Recognition sites for the following 17 restriction enzymes were used in the simulated digestions: AluI, BamHI, BfaI, BstUI (ThaI), DraI, EcoRI, HaeIII, HhaI, HinfI, HpaI, HpaII, KpnI, Sau3AI (MboI), MseI, RsaI, SspI, and TaqI. MW, ΦX174DNA digested by HaeIII.
The remaining 21 virtual RFLP patterns differed from the characteristic pattern of the previously described 16SrXII-A subgroup and shared similarity coefficients of 92–97%, confirming their affiliation with group 16SrXII and opening the possibility for recognition of new subgroups in this group. Thus, in accordance with criteria by Wei et al. (2007) and Lee et al. (2007), each of the 21 new RFLP patterns possibly identifies a new subgroup in group 16SrXII. However, while two of these RFLP patterns were repeatedly observed in cloned, amplified DNAs that were derived from independent samples and from independent PCRs, each of the 19 remaining RFLP patterns was observed only once. Because cloned PCR products were sequenced and PCR amplifications may introduce random errors (Chen et al., 1991), we choose to defer formal recognition of these 19 potentially new subgroups, describing them as tentative subgroups (16SrXII-A1 to 16SrXII-A19).
By contrast, the 16S rDNAs from phytoplasma strains BN-Op30 and BN-Op121 exhibited identical virtual RFLP patterns based upon digestions with 17 restriction enzymes. As the BstUI RFLP pattern distinguished strains BN-Op30 and BN-Op121 from strains in all previously described subgroups in group 16SrXII, these two strains are classified in new subgroup 16SrXII-F (XII-F) (Fig. 2 and Table 2[link]). The 16S rDNAs from phytoplasma strains BN-Op125 and BN-Fc3 exhibited identical virtual RFLP patterns, which distinguished (similarity coefficient ≤96%) these strains from strains in all previously described subgroups, including new subgroup XII-F, in group 16SrXII on the basis of digestion with AluI and BfaI (Fig. 2 and Table 2[link]). On the basis of these data, strains BN-Op125 and BN-Fc3 are placed in a new subgroup, XII-G. We note that actual gel electrophoresis RFLP analyses, carried out by using the distinguishing enzymes BstUI, AluI and BfaI on cloned F2nR2 PCR products from strains BN-Op30, BN-Op121, BN-Op125, and BN-Fc3 confirmed the virtual RFLP patterns (Fig. 3).
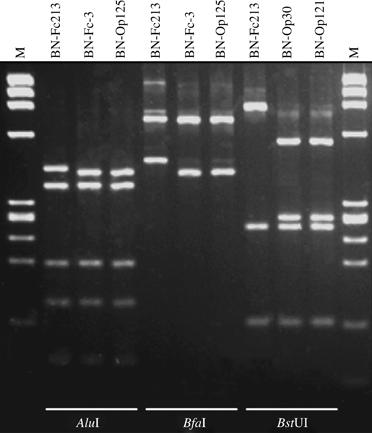
Actual gel-based polymerase chain reaction–restriction fragment length polymorphism profiles of F2nR2 amplicons from BN phytoplasma strains. Amplicons from Bois noir (BN)-Op30 and BN-Op121 (16SrXII-F), BN-Op125 and BN-Fc3 (16SrXII-G) and BN-Fc213 (16SrXII-A) were digested with AluI, BfaI and/or BstUI (ThaI), and the resultant restriction fragments were visualised by electrophoresis through 4% agarose gel. MW, ΦX174DNA digested by HaeIII.
Phylogenetic relationships
In BLAST searches, all the 79 cloned BN phytoplasma 16S rDNA sequences from the present study yielded best hits (matches) with phytoplasmas classified in subgroup 16SrXII-A (stolbur phytoplasma subgroup) (data not shown). A minimum evolution (ME) phylogenetic analysis of 16S rRNA gene sequences revealed that the 79 BN phytoplasma strains clustered together in a phylogenetic subclade with known phytoplasma strains of subgroup 16SrXII-A (data not shown). A phylogenetic tree based on 16S rDNA sequences from 58 previously characterised phytoplasma strains, 10 representative BN strains from this work, and Acholeplasma palmae is shown in Fig. 4.
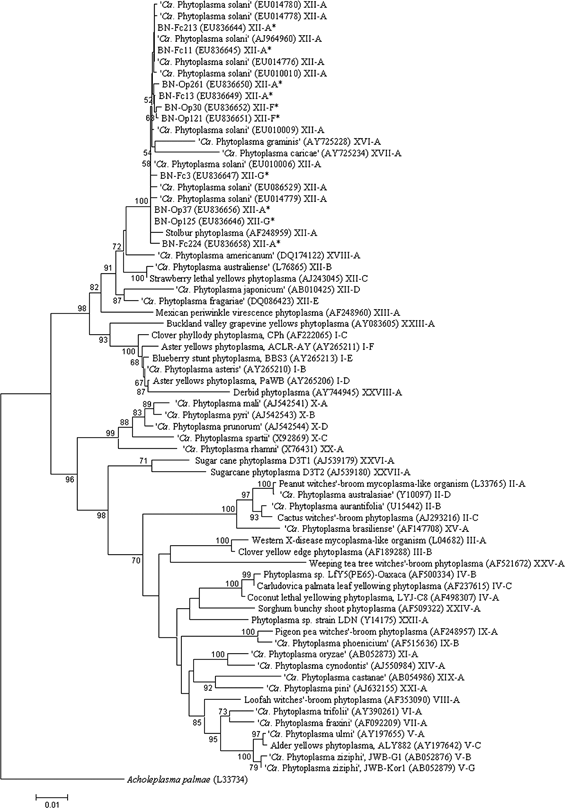
Phylogenetic tree inferred from phytoplasmal 16S rDNA F2nR2 fragments. Acholeplasma palmae was used to root the tree. Bootstrap values are displayed at tree nodes. GenBank accession numbers of nucleotide sequences are shown along with the names of phytoplasma strains. Nucleotide sequences determined in this study are indicated by asterisks.
Additional tentative new subgroups in group 16SrXII
In addition to the confirmed and tentative new group 16SrXII subgroups reported above, it is possible that additional subgroups may be represented by 16S rRNA gene sequences in public databases. For example, 16S rRNA gene sequences from six group 16SrXII phytoplasma strains of unknown subgroup affiliation were retrieved from the NCBI GenBank database and subjected to virtual RFLP analysis in this study. The 16S rDNAs from phytoplasma strains 400_05 (GenBank accession no. EU010007), 409_05 (GenBank accession no. EU010008) and 92_04 (GenBank accession no. EU014777), from grapevine plants in France and in Italy, exhibited mutually distinct (similarity coefficient ≤94%) virtual RFLP patterns that also differed (similarity coefficient ≤97%) from those characteristic of the seven confirmed and 19 tentative group XII subgroups (XII-A to XII-G and XII-A1 to XII-A19) (Table 2). These strains therefore may be tentatively considered as representative of three new subgroups 16SrXII-H, 16SrXII-I and 16SrXII-J, respectively.
The 16S rDNAs from phytoplasma strains 2005/2 from corn-reddening disease in Serbia (GenBank accession no. DQ222972), PorrocaC5 (GenBank accession no. EU131021) from Cocos nucifera in Panama, and RHOD1-CZ (GenBank accession no. DQ160245) from Rhododendron sp. in Czech Republic also exhibited mutually distinct (similarity coefficient ≤92%) virtual RFLP patterns that differed (similarity coefficient ≤97%) from those characteristic of the confirmed and tentative group XII subgroups (XII-A to XII-J and XII-A1 to XII-A19) (Table 2). Therefore, these strains may be tentatively considered as representative of three additional new subgroups 16SrXII-K, 16SrXII-L and 16SrXII-M, respectively.
Prior to this work, five subgroups had been described in the stolbur group (16SrXII): ‘Ca. Phytoplasma solani’ subgroup (16SrXII-A) (Firrao et al., 2005), ‘Ca. Phytoplasma australiense’ subgroup (16SrXII-B) (Davis et al., 1997), Strawberry lethal yellows phytoplasma subgroup (16SrXII-C) (Padovan et al., 2000), ‘Ca. Phytoplasma japonicum’ subgroup (16SrXII-D) (Sawayanagi et al., 1999) and ‘Ca. Phytoplasma fragariae’ subgroup (16SrXII-E) (Valiunas et al., 2006). The results of this study add two new, confirmed subgroups (XII-F and XII-G) as well as 22 tentative new subgroups (XII-A1 to XII-A19; XII-H, XII-I and XII-J) from phytoplasmas infecting grapevine and three tentative new subgroups (XII-K, XII-L and XII-M) from other hosts. Interestingly, the differences in restriction sites among the various subgroups is accounted for by SNPs at positions within restriction sites but other SNPs occur outside restriction sites tested in this study.
Additional 16S rDNA single nucleotide polymorphism lineages in Bois noir phytoplasma populations
Alignment of 79 1246-bp 16S rDNA sequences amplified, in PCRs primed by primer pair R16F2n/R16R2, from BN-diseased grapevine plants revealed SNPs at only three nucleotide positions (43 [T/C], 875 [A/G] and 1219 [T/G/A] respectively), none of which involved a recognition site for any of the 17 enzymes used to delineate 16SrXII subgroups in this study (Table 3). Of these, two (positions 43 and 1219) were located within recognition sites for two other restriction enzymes, MboII and FauI, respectively (Fig. 5a and Fig. 5b), although none of these three SNPs was located in a functional region discussed by Lafontaine & Tollervey (2001). SNPs at the three positions (43, 875 and 1219) are found among strains in subgroups XII-A, XII-F and XII-G. Nine 16Sr lineages, characterised by SNPs at positions 43, 875 and 1219 (Table 3), were each detected in at least three independent PCR products amplified from three separate diseased plants, supporting the conclusion that the SNPs were not because of errors in the PCRs. In addition, the SNP at nucleotide 875 is the same as that previously observed in SSCP analyses of 16S rDNA from ‘Ca. Phytoplasma solani’-related grapevine-infecting strains by Seruga Musićet al. (2008). The existence of the SNPs, within and outside of restriction endonuclease recognition sites in 16S rDNA, underscores the remarkable genetic diversity of ‘Ca. Phytoplasma solani’-related strains and affords molecular markers that should be useful for detailed investigation of the biology and epidemiology of grapevine BN and other stolbur phytoplasma-associated plant diseases.
| 16Sr lineage | Representative straina | Number of strainsb | 16Sr SNP positionc | Accession numberd | ||
|---|---|---|---|---|---|---|
| 43 | 875 | 1219 | ||||
| 16Sr-α | BN-Fc213 | 17 | T | A | T | EU836644 |
| 16Sr-β | BN-Fc11 | 5 | C | A | T | EU836645 |
| 16Sr-γ | BN-Op125 | 10 | T | G | T | EU836646 |
| 16Sr-δ | BN-Fc3 | 26 | C | G | T | EU836647 |
| 16Sr-ɛ | BN-Fc12 | 3 | C | A | G | EU836648 |
| 16Sr-ζ | BN-Fc13 | 3 | T | G | G | EU836649 |
| 16Sr-η | BN-Op261 | 4 | T | A | G | EU836650 |
| 16Sr-θ | BN-Op121 | 8 | C | G | G | EU836651 |
| 16Sr-ι | BN-Op30 | 3 | T | G | A | EU836652 |
- a Representative strain exhibiting the indicated 16Sr SNPs.
- b Total number of strains exhibiting the indicated 16Sr SNPs.
- c Bases from start of the F2nR2 fragment of 16S rRNA gene sequences.
- d GenBank accession number of 16S rRNA gene sequence from representative strain.
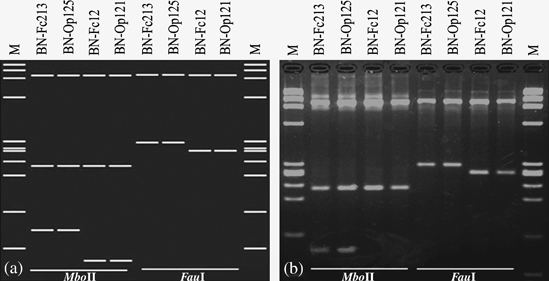
Virtual (a) and actual (b) MboII- and FauI-RFLP profiles from F2n/R2 amplicons. BN-Fc213 (lineage 16Sr-α), Bois noir-Franciacorta strain 213; BN-Op125 (lineage 16Sr-δ), Bois noir-Oltrepo’ pavese strain 125; BN-Fc12 (lineage 16Sr-ɛ), Bois noir-Franciacorta strain 12; BN-Op121 (lineage 16Sr-θ), Bois noir-Oltrepo’ pavese strain 121; M, molecular marker Φx174 digested by HaeIII.
Genetic diversity and ecology of stolbur phytoplasma strains
Identification of SNPs delineating no less than two new grapevine-associated 16SrXII subgroups and of other SNPs in the present study opens new opportunities for in-depth studies of the distribution of grapevine-associated 16SrXII phytoplasma strains in weeds, insect vector populations and grapevines from vineyards located in different geographic areas. Application of automated virtual restriction analysis should facilitate such studies as in other work. For example, on the basis of automated virtual RFLP analysis of 16S rDNA, genetic diversity among cactus witches’ broom (CaWB) phytoplasma strains infecting Opuntia in China was correlated with environmental conditions in cactus growing regions (Cai et al., 2008). It has been reported that weeds host various stolbur phytoplasma strains (Langer & Maixner, 2004) and that H. obsoletus preferentially feeds on certain plant species in different geographic areas (Alma et al., 1987; Sforza et al., 1999; Langer & Maixner, 2004; Sharon et al., 2005; Lessio et al., 2007). It would be interesting to learn whether particular SNP lineage(s) could be correlated with BN symptom severity, efficiency and specificity of transmission by vectors, and regional frequency of BN in vineyards of Europe.
Acknowledgements
This study was supported by the project ‘GIAVI’ of MiPAF (Italian Ministry of Agriculture) and the Project REFLAVI (Regione Lombardia). We thank Prof. Giuseppe Belli for critically reading the manuscript.




