Multigene analysis for differentiation of aster yellows phytoplasmas infecting carrots in Serbia
Abstract
During a survey of large carrot fields in Serbia, plants showing leaf reddening and/or yellowing, adventitious shoot production and reduction in taproot size and quality were observed in a low percentage of plants. To verify phytoplasma association with the described symptoms and to carry out pathogen differentiation, PCR assays followed by restriction fragment length polymorphism (RFLP) analyses and/or sequencing of phytoplasma 16Sr DNA and ribosomal protein genes l22 and s3, tuf, putative aa kinase plus ribosomal recycling factor genes and DNA helicase gene were carried out. Phytoplasmas belonging to 16SrI-A and 16SrI-B ribosomal subgroups and to rpI-A and rpI-B ribosomal protein subgroups, respectively, were identified by RFLP analyses in 13 of 15 symptomatic plants tested. No amplification was obtained with non-symptomatic carrot samples. The identification was confirmed by sequence analyses of the phytoplasma genes studied. In two carrot samples, presence of interoperon sequence heterogeneity was detected and phytoplasma strains were identified as belonging to 16SrI group but were not assigned to any 16S rRNA or ribosomal protein subgroup. This research allowed the first molecular identification of phytoplasmas infecting carrot in Serbia using several molecular markers, and it indicates that under field conditions in non-epidemic outbreaks a certain amount of genetic mutation may occur in conserved genes of these prokaryotes.
Introduction
‘Candidatus Phytoplasma asteris’ (aster yellows, AY) (Lee et al., 2004), beet leafhopper-transmitted virescence agent (BLTVA) and Spiroplasma citri (SC) are mollicutes reported to infect carrot (Daucus carota L.) worldwide, causing indistinguishable symptoms on infected plants (Lee et al., 2006a). All these pathogens are prokaryotes with phloematic habitats that can be detected both in wild and in cultivated plants and are transmitted by leafhoppers that may have occasional or permanent trophic relationship with their hosts.
During a survey of large carrot fields at Begeč, Bačka region, Serbia, symptoms referable to phytoplasma infection were observed. The symptoms resembled those already described in carrot in North America and in Israel (Orenstein et al., 1999; Lee et al., 2003, 2006a) that had been associated with the phytopathogenic mollicutes described above.
In a preliminary work, we have determined that aster yellows phytoplasmas are associated with this disease (Duduk et al., 2007). In the present study, we investigated the genetic variability of aster yellows phytoplasmas detected in carrot fields under non-epidemic conditions. The current aster yellows phytoplasma classification, as the molecular phytoplasma classification in general, relies on PCR amplification of 16S rDNA followed by restriction fragment length polymorphism (RFLP) analysis and/or sequencing (IRPCM, 2004). However, this approach does not always provide a clear molecular distinction, and because the aster yellows group encompass phytoplasmas infecting numerous different plant species and insect vectors (Lee et al., 2004), it is not helpful for epidemiological studies towards possible control of these diseases. Therefore, in this study, four additional genes to differentiate the aster yellows phytoplasma strains detected in carrot samples were employed. These strains were compared with reference strains previously reported to be associated with carrot and other herbaceous hosts.
Materials and methods
Sample collection and nucleic acid extraction
Samples were collected from 15 symptomatic and 2 asymptomatic carrot plants in the South Bačka region of Serbia, during October 2006. Total nucleic acids were extracted from 0.5 g of fresh leaf tissue following the protocol described by Angelini et al. (2001), dissolved in TE buffer and maintained at −20°C. Nucleic acids were quantified and diluted in sterile distilled water to the final concentration of 20 ng μL−1 before performing PCR assays.
Phytoplasma strains
The phytoplasma strains Chrysanthemum yellows (CHRY, ribosomal subgroup 16SrI-A), European aster yellows (EAY, ribosomal subgroup 16SrI-B), Catharanthus virescence (CVB, ribosomal subgroup 16SrI-F), carrot yellows (CA, ribosomal group 16SrI-C), primula yellows (PRIVA, ribosomal subgroup 16rI-L), clover phyllody from France (KVF, ribosomal subgroup 16SrI-C) and stolbur from pepper from Serbia (STOL C, ribosomal subgroup 16SrXII-A), maintained in collection in periwinkle [Catharanthus roseus (G.) Don.] (Bertaccini, 2003), were employed as reference strains in RFLP analyses.
Polymerase chain reaction amplification and sequence analyses
16S ribosomal DNA
Direct PCR assays with the universal phytoplasma primer pair P1/P7 (Deng & Hiruki, 1991; Schneider et al., 1995) or with primer pair R16(I)F1/R1 (Lee et al., 1994) specific to the ribosomal groups I, II (Tolu et al., 2006) and XII were carried out for phytoplasma detection. Each 25 μL PCR reaction mix contained 20 ng template DNA, 2.5 μL 10× PCR buffer, 0.8 U Taq polymerase (Polymed, Florence, Italy), 0.2 mM dNTPs, 1.5 mM MgCl2 and 0.4 mM of each primer. Samples lacking DNA were employed as negative controls. Thirty-five PCR cycles were performed under the following conditions: 1 min (2 min for the first cycle) for denaturation step at 94°C, 2 min for annealing at 50°C and 3 min (10 min for the last cycle) for primer extension at 72°C. Six microlitres of PCR products were separated in 1% agarose gel, stained with ethidium bromide and visualised with UV transilluminator.
Identification of detected phytoplasmas was performed using RFLP analyses with Tsp509I (New England Biolabs, Beverly, MA, USA), TaqI, HhaI and TruI (Fermentas, Vilnius, Lithuania) restriction enzymes. RFLP products were separated in a 5% polyacrylamide gel, stained with ethidium bromide and visualised under UV transilluminator.
The P1/P7-amplified products of carrot samples 2006/1, 2006/5 and 2006/9 were purified using Qiagen PCR purification kit (Qiagen GmbH, Hilden, Germany) and sequenced in both directions with two forward primers P1 and R16F2 (Lee et al., 1995) and one reverse primer P7, using the BIG DYE sequencing terminator kit (PE Biosystems, Warrington, UK). A database search of homologous sequences was performed by Blast analyses at the National Center for Biotechnology Information (NCBI) website (http://ncbi.nlm.nih.gov/BLAST).
Ribosomal protein genes
A further molecular characterisation was performed by PCR using rpF1C/rp(I)R1A primer pair (Martini et al., 2007) that amplifies part of the ribosomal operon, which includes the 3′ end of the s19 gene and the complete l22 and s3 genes (Lim & Sears, 1992). The PCR reaction mix and negative control were as described above. Thirty-eight PCR cycles were performed under the following conditions: 1 min (2 min for the first cycle) for denaturation step at 94°C, 2 min for annealing at 50°C and 3 min (10 min for the last cycle) for primer extension at 72°C. PCR product separation, RFLP analyses using TruI, Tsp509I, AluI and DdeI (Fermentas) and visualisation of PCR and RFLP products were performed as described above.
The rpF1C/rp(I)R1A-amplified products of carrot samples 2006/1, 2006/5 and 2006/9 were purified, sequenced with primers rpF1C and rp(I)R1A and deposited in the NCBI (Bethesda, MD, USA) as described above. These sequences were aligned with those of 24 representative ‘Ca. P. asteris’ accessions available in GenBank (Table 2) using CLUSTALX program (Thompson et al., 1997) and BioEdit (Hall, 1999). A phylogenetic tree was constructed using MEGA version 4 (Tamura et al., 2007). Acholeplasma laidlawii, a cultivable mollicute, phylogenetically related to phytoplasmas, was designated as the outgroup to root the tree. A database search of homologous sequences was also performed as described above.
| Phytoplasma strain | 16Sr group | rp group | Accession number | References | |
|---|---|---|---|---|---|
| 16S rDNA | rp genes | ||||
| 2006/1 | I-A | I-A | EU215424 | EU215428 | This study |
| 2006/5 | n.d. | n.d. | EU215425 | EU215429 | This study |
| 2006/9 | I-B | I-B | EU215426 | EU215430 | This study |
| Br273 | I-B | I-B | EU215427 | EU215431 | This study |
| Btsv2CarD1 | I-A | I-A | AY180926.1 | AY183690 | Lee et al. (2003) |
| Btsv2CarD3 | I-B | I-B | AY180945.1 | AY183710 | Lee et al. (2003) |
| CabD3 | I-B | I-B | AY180947.1 | AY183717 | Lee et al. (2003) |
| OnionD2 | I-A | I-A | AY180931.1 | AY183699 | Lee et al. (2003) |
| ParsD1 | I-B | I-B | AY180954.1 | AY183719 | Lee et al. (2003) |
| ParsD3 | I-A | I-A | AY180940.1 | AY183700 | Lee et al. (2003) |
| PLD1 | I-A | I-A | AY180941.1 | AY183702 | Lee et al. (2003) |
| RgwdD1 | I-A | I-A | AY180930.1 | AY184704 | Lee et al. (2003) |
| Btsv2M.f.12 | I-B | I-B | AY180951.1 | AY183711 | Lee et al. (2003) |
| BtsvS.i.4 | I-A | I-A | AY180938.1 | AY183695 | Lee et al. (2003) |
| Btsv2C.a.13 | I-A | I-A | AY180925.1 | AY183688 | Lee et al. (2003) |
| Btsv2C.a.17 | I-B | I-B | AY180944.1 | AY183716 | Lee et al. (2003) |
| AV2192 | I-L | I-B | AY180957.1 | AY183708 | Lee et al. (2003) |
| BB | I-A | I-A | AY180955.1 | AY183686 | Lee et al. (2003) |
| CHRY | I-A | I-A | AY180956.1 | AY183696 | Lee et al. (2003) |
| MIAY | I-B | I-B | M30790.1 | M74770 | Lee et al. (2003) |
| CVB | I-F | I-N | AY265212.1 | AY264865 | Martini et al. (2007) |
| MBS | I-B | I-L | AY265208.1 | AY264858.1 | Lee et al. (2004) |
| PaWB | I-D | I-D | AY265206.1 | AY264857.1 | Lee et al. (2004) |
| IoWB | I-N | I-F | AY65205.1 | AY264859.1 | Lee et al. (2004) |
| BBS3 | I-E | I-E | AY265213.1 | AY264863.1 | Lee et al. (2004) |
| STRAWB2 | I-K | I-J | U96616 | U96617.1 | Lee et al. (2004) |
| CPh | I-C | I-C | AF222065 | AY264862.1 | Lee et al. (2004) |
| GD1 | I-A | I-M | DQ112021 | AY264864.1 | Lee et al. (2006b) |
| A. ladlawii | n.a. | n.a. | M23932 | M74771 | |
- n.a., group not available; n.d., group not defined.
Other chromosomal DNA fragments
Molecular analyses were carried out using the tuf gene coding the elongation factor Tu and the putative aa kinase gene and ribosomal recycling factor gene (Botti & Bertaccini, 2003) amplified using TufAYf/r (Schneider et al., 1997) and BB88F1/R1 (Gundersen et al., 1996) primer pairs, respectively. The PCR reaction mix and negative control were as described above. For both primer pairs, 35 PCR cycles were performed under the following conditions: 30 s for denaturation step at 95°C, 30 s for annealing at 55°C and 1 min for primer extension at 72°C. PCR products were visualised as described above. RFLP analysis was performed using Tsp509I and TruI restriction enzymes on BB88F1/R1 and only TruI restriction enzyme on tuf gene amplicons.
The G35p/m primer pair was used with parameters described by Davis et al. (1992) to amplify a phytoplasma DNA helicase gene (Duduk & Bertaccini, 2006). RFLP analyses were carried out using TruI and AluI restriction enzymes. Visualisation of RFLP products was performed as described above.
Results
Symptoms
The affected plants showed reddening, purpling and yellowing of the leaves, formation of chlorotic adventitious shoots and reduction in the size and quality of taproots (Fig. 1).

Symptomatic carrot (A) showing leaf reddening, formation of chlorotic adventitious shoots and reduction in taproot size and quality, and asymptomatic carrot (B).
Polymerase chain reaction amplification and sequence analyses
16S ribosomal DNA
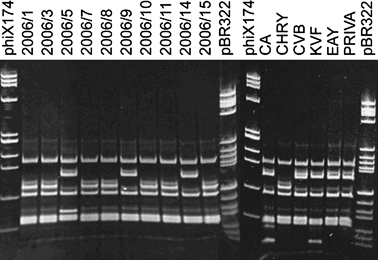
Polymerase chain reaction reactions with P1/P7 and R16(I)F1/R1 primer pairs resulted in amplification of the expected fragment length of about 1800 and 1100 bp, respectively, from all the 15 symptomatic carrot samples tested. No amplification was obtained from the two asymptomatic samples tested. The restriction profiles obtained with amplicons P1/P7 using HhaI restriction enzyme (Fig. 2 and data not shown), as well as with R16(I)F1/R1 amplicons using HhaI and TruI restriction enzymes allowed two different groups of profiles to be distinguished (Table 1); one of them with both amplicons was indistinguishable from the reference strain CHRY, which belongs to ribosomal subgroup 16SrI-A, while the other profile was indistinguishable from the reference strain EAY, belonging to ribosomal subgroup 16SrI-B. However, the restriction profiles obtained with amplicons P1/P7 using Tsp509I restriction enzyme (Fig. 2) allowed three different groups of profiles to be distinguished (Table 1). The profiles of 13 samples were consistent with the two described above, while those of samples 2006/5 and 2006/6 were not identical to any employed reference strain. The restriction profiles of the same 13 samples using TaqI restriction enzyme (Fig. 2) showed no polymorphism (Table 1), while the profiles of samples 2006/5 and 2006/6 were again different from the others and from all phytoplasma controls employed. The restriction profiles obtained with the same amplicons from all carrot samples using TruI restriction enzyme showed no polymorphism (Table 1).
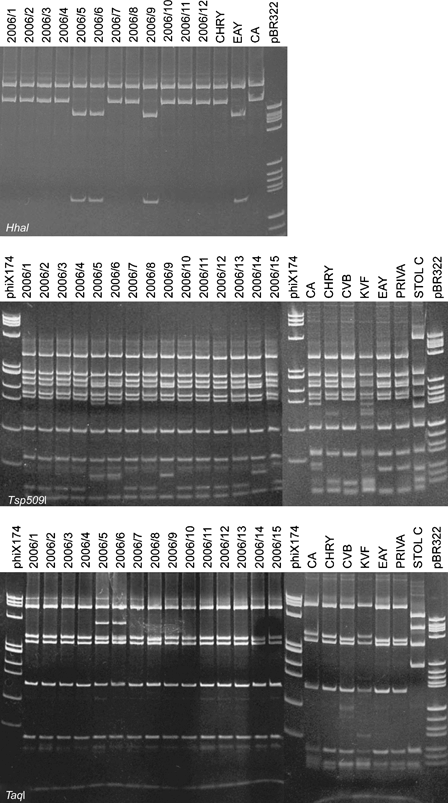
Polyacrylamide gel 5% showing the HhaI, Tsp509I and TaqI restriction fragment length polymorphism patterns of phytoplasma 16S rDNA plus spacer region amplified with P1/P7 primer pair from carrot samples and from phytoplasma reference strains in periwinkle. Sample abbreviations: 2006/1–2006/15, carrot samples; CA, carrot yellows (16SrI-C); CHRY, Chrysanthemum yellows (16SrI-A); CVB, Catharanthus virescence (16SrI-F); KVF, clover phyllody from France (16SrI-C); EAY, European aster yellows (16SrI-B); PRIVA, primula yellows (16SrI-L); STOL C, stolbur from pepper from Serbia (16SrXII-A); phiX174, marker phiX174 HaeIII digested; fragment sizes in base pairs from top to bottom: 1353, 1078, 872, 603, 310, 281, 271, 234, 194, 118 and 72; pBR322, marker pBR322 HaeIII digested; fragment sizes in base pairs from top to bottom: 587, 540, 502, 458, 434, 267, 234, 213, 192, 184, 124, 123, 104, 89, 80, 64, 57 and 51.
| Gene | 16S Ribosomal in R16(I)F1/R1 plus Spacer Region (P1/P7) | Ribosomal Protein l22 and s3 | tuf | aak + rrf | DNA Helicase | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Primers | P1/P7 | R16(I)F1/R1 | rpF1C/rp(I)R1A | TufAYf/r | BB88F1/R1 | G35p/m | ||||||
| Straina | HhaI | TruI | Tsp509I | TaqI | HhaI, Tsp509I | AluI | Tsp509I, TruI | DdeI | TruI, Tsp509I | Tsp509I | TruI | TruI, AluI |
| 2006/1 | A | A | A | A | A | A | A | A | A | A | A | A |
| 2006/2 | A | A | A | A | A | A | A | A | A | A | A | A |
| 2006/3 | A | A | A | A | A | A | A | A | A | A | A | — |
| 2006/4 | A | A | A | A | A | A | A | A | A | A | A | — |
| 2006/5 | B | A | C | B | B | B | C | A | B | B | C | — |
| 2006/6 | B | A | C | B | B | B | C | A | B | B | C | — |
| 2006/7 | A | A | A | A | A | A | A | A | A | A | A | A |
| 2006/8 | A | A | A | A | A | A | A | A | A | A | A | A |
| 2006/9 | B | A | B | A | B | B | B | A | B | B | B | A |
| 2006/10 | A | A | A | A | A | A | A | A | A | A | A | A |
| 2006/11 | A | A | A | A | A | A | A | A | A | A | A | A |
| 2006/12 | A | A | A | A | A | A | A | A | A | A | A | A |
| 2006/13 | A | A | A | A | A | A | A | A | A | A | A | A |
| 2006/14 | B | A | B | A | B | B | B | A | B | B | B | — |
| 2006/15 | A | A | A | A | A | A | A | A | A | A | A | — |
- A, B, C, distinct restriction fragment length polymorphism profiles; aaK, aminoacid kinase; rrf, ribosomal recycling factor.
- a Strains sequenced are given in bold.
The sequences obtained from carrot samples 2006/1, 2006/5 and 2006/9 (1736, 1708 and 1720 bp, respectively) were deposited in the NCBI under accession numbers EU215424, EU215425 and EU215426. The DNA sequence chromatogram of sample 2006/5 clearly indicated six ambiguous bases, and this sequence was therefore excluded from further phylogenetic analyses.
The 16Sr DNA plus spacer sequence of sample 2006/1 showed the highest identity value of 99% with aster yellows phytoplasma strains from Lithuania and Ohio (USA) (AF510323.1 and CP000061.1, respectively), while the same region of sample 2006/9 showed the highest identity value of 99% with aster yellows phytoplasma strains from Hawaii (USA), Lithuania and Japan (AY665676.1, AY102274.1 and AP006628.1, respectively).
A similar comparison of the 16S rRNA gene alone of samples 2006/1 and 2006/9 with 24 strains of aster yellows phytoplasmas (Table 2) confirmed that the phytoplasma detected in carrot sample 2006/1 was most closely related to aster yellows phytoplasma reference strains belonging to the 16SrI-A ribosomal subgroup, while the phytoplasma detected in carrot sample 2006/9 was closely related to phytoplasmas of the 16SrI-B ribosomal subgroup as defined by Lee et al. (1998a,b)(data not shown).
Ribosomal protein genes
The sequences obtained from samples 2006/1, 2006/5 and 2006/9 (1137, 1123 and 1132 bp, respectively) were deposited in the NCBI, under accession numbers EU215428, EU215429 and EU215430.
The PCR assays with rpF1C/rp(I)R1A primer pair resulted in amplification of the expected fragment length of about 1200 bp from all symptomatic carrot samples tested. RFLP analyses with TruI and AluI restriction enzymes (Fig. 3) produced two different restriction profiles, one of which was indistinguishable from that of reference strain CHRY (16SrI-A), while the other was indistinguishable from that of reference strain EAY (16SrI-B) (Fig. 3). RFLP analyses of the same amplicons with DdeI restriction enzyme showed no polymorphism (Table 1). The RFLP analyses with Tsp509I restriction enzyme produced three different restriction profiles, one of which was indistinguishable from that of reference strain CHRY, the second was indistinguishable from that of reference strain EAY, while the third did not match with any profile of 16SrI reference phytoplasmas employed (Fig. 3; Table 1).
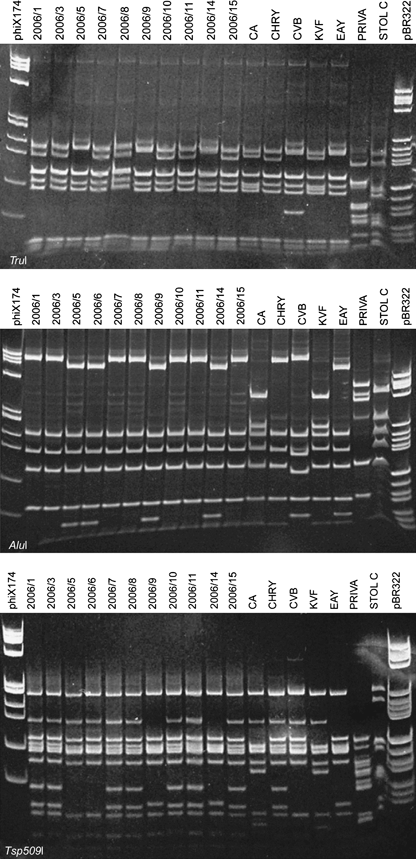
Polyacrylamide gel 5% showing the TruI, AluI and Tsp509I restriction fragment length polymorphism patterns of phytoplasma l22 and s3 genes amplified with rpF1C/rp(I)R1A primer pair from carrot samples and from phytoplasma reference strains in periwinkle. Sample abbreviations: 2006/1–2006/15, carrot samples; see Fig. 2 for abbreviations of reference strains; phiX174, marker phiX174 HaeIII digested; fragment sizes in base pairs from top to bottom: 1353, 1078, 872, 603, 310, 281, 271, 234, 194, 118 and 72; pBR322, marker pBR322 HaeIII digested; fragment sizes in base pairs from top to bottom: 587, 540, 502, 458, 434, 267, 234, 213, 192, 184, 124, 123, 104, 89, 80, 64, 57 and 51.
The 3′ end of the s19 gene and the complete l22 and s3 genes of sample 2006/1 was identical with aster yellows phytoplasma strains from Chrysanthemum (AY264869.1) and Plantago coronopus from Germany (AY264867.1), and with false ragweed (AY183705.1 and AY183704.1), prickly lettuce (AY183702.1 and AY183697.1), carrot (AY183694.1, AY183692.1 and AY183691.1), Ceratagallia abrupta (AY183693.1, AY183689.1 and AY183688.1) and tomato from Texas (AY183686.1) and lettuce (CP000061.1). However, the sequence of the same amplicon from sample 2006/5 showed the highest identity value of 99% with maize bushy stunt (MBS) phytoplasma strain from Mexico (AY264858), while the same region of sample 2006/9 was identical with Texas aster yellows phytoplasma strains from cabbage (AY183717.1), Ceratagallia abrupta (AY183716.1), Scaphytopius irroratus (AY183715.1, AY183712.1), Macrosteles fascifrons (AY183711.1) and carrot (AY183710.1).
Phylogenetic comparison of the l22 and s3 genes of samples 2006/1, 2006/5 and 2006/9 with 24 representative strains of phytoplasmas from aster yellows ribosomal group indicated that the phytoplasma detected in carrot samples 2006/1 and 2006/9 can be enclosed respectively in the rpI-A and rpI-B ribosomal protein subgroups as defined by Lee et al. (1998a,b), while sample 2006/5 was found to be very closely related to MBS phytoplasma, which belong to rpI-L ribosomal protein subgroup (Lee et al., 2004) (Fig. 4). Aligned putative restriction site maps for Hpy8I and HpyCH4III restriction enzymes of carrot samples 2006/1, 2006/5 and 2006/9 and of phytoplasma strain MBS revealed characteristic restriction profiles differentiating among these strains (data not shown).

Phylogenetic tree constructed using neighbour-joining algorithm of ribosomal protein operon sequences from carrot samples from Serbia, 1 broccoli sample from Serbia, 11 samples from Lee et al. (2003) and 12 reference strains, employing Acholeplasma laidlawii as the outgroup. Phytoplasma strains are described in Table 2.
Other chromosomal DNA fragments
Polymerase chain reaction with the primers TufAYf/r and BB88F1/R1 resulted in amplification of the expected fragment length of about 1200 and 740 bp, respectively, from all symptomatic carrot samples tested. RFLP analyses with TruI restriction enzyme on TufAYf/r amplicons and with Tsp509I restriction enzyme on both amplicons provided two different groups of profiles (Table 1). One group of profiles with both amplicons was indistinguishable from strain CHRY, while the other group was indistinguishable from strain EAY. RFLP analyses with TruI restriction enzyme on BB88F1/R1 amplicon produced three different restriction profiles, one of which was indistinguishable from the profile of reference strain CHRY, the second was indistinguishable from reference strain EAY, while the third did not match any reference phytoplasma profile (Fig. 5).
Polyacrylamide gel 5% showing the TruI restriction fragment length polymorphism patterns of phytoplasma putative aa kinase gene and ribosomal recycling factor amplified with BB88F1/R1 primer pair from carrot samples. Sample abbreviations: 2006/1–2006/15, carrot samples; see Fig. 2 for abbreviations of reference strains; phiX174, marker phiX174 HaeIII digested; fragment sizes in base pairs from top to bottom: 1353, 1078, 872, 603, 310, 281, 271, 234, 194, 118 and 72. pBR322, marker pBR322 HaeIII digested; fragment sizes in base pairs from top to bottom: 587, 540, 502, 458, 434, 267, 234, 213, 192, 184, 124, 123, 104, 89, 80, 64, 57 and 51.
Polymerase chain reaction with G35p/m primer pair resulted in amplification of the expected fragment length of about 1200 bp only from 9 carrots of 15 symptomatic samples tested that showed no polymorphisms in RFLP analyses with TruI and AluI restriction enzymes (Table 1).
Discussion
The obtained results confirmed that the symptoms observed in carrot plants are associated with the presence of aster yellows phytoplasmas (Ca P. asteris), in particular, with strains belonging to 16SrI-A and 16SrI-B subgroups. Phytoplasmas belonging to the same 16SrI subgroups had also been identified together with BLTVA and Spiroplasma citri (SC) in North American carrot fields where plants with symptoms similar to those observed in Serbia were described (Lee et al., 2006a).
This study presents the first molecular identification of phytoplasmas in carrot in Serbia obtained with different molecular markers to characterise phytoplasma population infecting carrots in the field. Based on the RFLP analyses on 16S ribosomal RNA gene, spacer region and beginning of 23S ribosomal gene (P1/P7 amplicons), two carrot samples (2006/5 and 2006/6) were identified as members of group 16SrI, but collective RFLP profiles differed from those of 16SrI reference strains. The DNA fragments obtained with TaqI restriction enzyme were approximately 0.8, 0.5, 0.4, 0.35 and 0.15 kbp in size, which is in total larger than P1/P7 expected PCR products (approximately 1.7 kbp). That leads to a hypothesis that those profiles represent two differentiable ribosomal operons, and this hypothesis is supported by the DNA sequence chromatogram indicating the presence of probable point mutations. The possibility that two phytoplasma strains are present in a sample as a mixed infection is unlikely because the other analysed genes clearly indicated the presence of a single sequence with unique RFLP pattern. The presence of 16S rRNA interoperon sequence heterogeneity is not uncommon and has been reported in 16SrI and several other ribosomal groups (Schneider & Seemüller, 1994; Liefting et al., 1996; Lee et al., 1998a; Jomantiene et al., 2002; Davis et al., 2003). Although the difference in sequence homology between two operons is relatively small (0.2–0.7%), when differences occur in restriction sites, misidentification or erroneous assignment of the same phytoplasma to two different 16S rRNA subgroups is possible because classification is mainly based on the RFLP analyses of 16S rDNA (Davis et al., 2003; IRPCM, 2004). In those cases, cloning of PCR products and analyses of both rRNA operons separately can help in understanding the problem (Davis et al., 2003); however, the use of other genes present as single copy in the phytoplasma genome can discriminate when different phytoplasma populations are present in mixed infection from interoperon sequence heterogeneity.
Based on the RFLP analyses with Tsp509I restriction enzyme on ribosomal protein genes l22 and s3 [rpF1C/rp(I)R1A amplicons], all phytoplasmas within the subgroup 16SrI-A were classified as rp subgroup rpI-A, all phytoplasmas within the subgroup 16SrI-B were classified as rp subgroup rpI-B, while the two phytoplasmas showing interoperon heterogeneity were not classified at rpI subgroup level (Lee et al., 2003; Martini et al., 2007). The RFLP analyses with TruI restriction enzyme on the putative aa kinase gene and ribosomal recycling factor (BB88F1/R1 amplicon) confirmed that these two phytoplasmas, in these genes, are also different from those already described as members of aster yellows group. These results confirm the importance of multiple gene analyses for differentiation among field-collected phytoplasma strains and are in agreement with results reported for 16SrI group phytoplasmas in periwinkle samples artificially infected (Botti & Bertaccini, 2003).
The RFLP analyses of tuf gene (TufAYf/r amplicon) is in agreement with the results obtained with 16Sr DNA, ribosomal protein genes and BB88F1/R1 amplicons showing no difference among the 11 strains from carrot identified as belonging to 16SrI-A ribosomal subgroup and CHRY reference phytoplasma strain representing 16SrI-A ribosomal subgroup. On the contrary, the RFLP analyses on tuf gene showed no difference among the strains classified as 16SrI-B, the two strains not classified at the subgroup level and EAY reference phytoplasma strain representing 16SrI-B ribosomal subgroup.
Sequence homology, phylogenetic and virtual restriction analyses of ribosomal protein genes confirmed that 16SrI-A rpI-A and 16SrI-B rpI-B phytoplasmas from carrot samples cluster together with the strains described in the same ribosomal and ribosomal protein subgroup, respectively, while the previously unreported strain shows major homology to MBS phytoplasma from which it can however be distinguished. These results are consistent with RFLP and virtual RFLP analyses, confirming that this strain is genetically close to 16SrI-B and rpI-L subgroups but distinct from both.
Aster yellows phytoplasmas have been described to infect a wide variety of host plants worldwide, and studies of 16Sr DNA alone in some cases of interoperon heterogeneity are not sufficient or suitable to resolve molecular differences among strains. In our case, data obtained with RFLP and sequence analyses of less conserved genes substantiated those obtained through RFLP and sequence analyses of rDNA (16S rRNA gene and 16S/23S rDNA spacer region) but did not allow a more detailed differentiation of the Serbian phytoplasma strains infecting carrot. However, because interoperon sequence heterogeneity is present, the clear molecular differentiation and definition of these strains was achieved by studying other less conserved genes such as ribosomal protein or putative aa kinase gene and ribosomal recycling factor. These differences can be relevant for epidemiological studies that are the basis for controlling the spreading of phytoplasma diseases in the fields. The identification of a new phytoplasma strain in carrot fields in which infection was high, but not at epidemic level, indicates that a certain amount of genetic mutation can occur under such conditions, leading to appearance of new strains with possibly different epidemic capacities. The use of more genes, in addition to 16Sr DNA, for phytoplasma classification was shown to be relevant for studying the epidemiology of other phytoplasma diseases such as ‘flavescence dorée’ in which the presence of phytoplasma strains, differentiated on the basis of several genetic markers, was proved to be useful for determining their epidemic ability (Martini et al., 2002; Botti & Bertaccini, 2007, Arnaud et al., 2007). It is clear from the results presented here that more genes beside 16Sr DNA should be applied for phytoplasma identification, as it has already been suggested by other authors (Liefting et al., 1996; Lee et al., 2006b). For aster yellows phytoplasmas, the rp and aa kinase genes plus ribosomal recycling factor can be used for this purpose.




