Analysis of genetic diversity and phylogenetic relationship of red deer subspecies in XinJiang, China
ABSTRACT
Polymorphisms for seven microsatellite loci in three red deer subspecies (9 populations) found in XinJiang were detected by polymerase chain reaction (PCR), 12% nondenaturation polyacrylamide gel electrophoresis and the Sanguinetti silver staining method. Numbers of alleles, average effective numbers of alleles (E) and the average rate of homozygosity, allelic frequencies of seven microsatellite loci, polymorphism information content (PIC), mean heterozygosity (H) and genetic distances among the populations were calculated for each population. Dendrograms were constructed based on genetic distances by the neighbor-joining method (NJ), utilizing molecular evolutionary genetics analysis software PHYLIP (3.6). The phylogenetic tree was constructed based on allelic frequencies using maximum likelihood (ML); the bootstrap value was estimated by bootstrap test in the tree. Lastly, phylogenesis was analyzed. The results showed that four of the seven microsatellite loci were highly polymorphic, but BMS2508 and Celjp0023 showed no polymorphism and BM5004 was a neutral polymorphism. It is our conclusion that the four microsatellite loci are effective DNA markers for the analysis of genetic diversity and phylogenetic relationships among the three red deer subspecies. The mean PIC, H and E-values across the microsatellite loci were 0.5393, 0.5736 and 2.64, which showed that these microsatellite loci are effective DNA markers for the genetic analysis of red deer. C.e. songaricus populations from Regiment 104, 151 and Hami are clustered together. C.e. yarkandensis populations from Regiment 35, Xaya and Alaer are clustered together. These two clusters also cluster together. Lastly, C.e. sibiricus populations from Burqin, Regiment 188 and the first two clusters were clustered together. The phylogenetic relationship among different red deer populations is consistent with the known origin, history of breeding and geographic distributions of populations.
INTRODUCTION
Microsatellite loci are molecular markers based on 1–6 bp short tandem repeats, and they are extensively distributed throughout the genome. Microsatellite loci have been used for the study of genetic diversity, genetic distance and dendrograms, due to their abundance, high polymorphism throughout the genome, rapid and convenient detection, and they are subjected to less selective pressure than other genomic sequences(Hamada et al. 1982; Tautz 1989; Takezaki & Nei 1996). The number of red deer has grown in XinJiang because of effective conservation measures. Study of the genetic diversity and genetic relationships among red deer subpopulations in XinJiang will assist with the development of effective measures to further protect and exploit them, as well as contribute to the study of the origin and evolution of the province's red deer populations.
Randi et al. (1998, 2001) analyzed the complete sequence of mitochondrial cytochrome b (Cytb) gene in red deer subspecies in Europe, Asia and North America. Their results showed that the diversity of the complete sequence of the Cytb gene was 0.09% to 1.75% in six red deer subspecies in China. This level of diversity suggests that the variation between Chinese red deer breeds is at the subspecies level. Li et al. (1998) also studied the origin and evolution of the complete sequence of the Cytb gene of red deer subspecies, and indicated that the DNA differences of four subspecies were within subspecific diversity. De Young et al. (2003) studied the population structure of Ozotoceros bezoarticus deer by analysis of the control region of mtDNA. Their results seemed to confirm the importance of rapid population expansion and habitat continuity in retaining genetic variation in restored populations. However, the use of diverse transplanted stocks and the varied demographic histories of populations resulted in fine-scale genetic structuring. There are a few studies of the red deer subspecies in XinJiang (Mahmut et al. 2001). In the present study, genetic diversity and the evolutionary relationship among three red deer subspecies in XinJiang province (Ainiwaer et al. 2008) were examined utilizing seven microsatellite loci DNA markers.
MATERIALS AND METHODS
Sample collection
Samples for DNA analysis were collected from the central producing area in XinJiang. Breed names, sample size and localities of the populations studied are listed in Table 1. Six milliliters of blood were taken from the cervical vein into a tube with anticoagulant, and stored in at −20°C in a freezer until analysis.
| Populations | No. of individuals | Total No. in each farm | Localities (abbreviation) |
|---|---|---|---|
| C.e. songaricus | 33 | 812 | Hong Shan deer farm in HAMI (TSHM) |
| C.e. songaricus | 32 | 655 | Deer farm in REGIMENT 104 (TS104) |
| C.e. songaricus | 20 | 221 | Deer farm in REGIMENT 151 (TS151) |
| C.e. songaricus Wild | 32 | 257 | Tianshan forest in QITAI (TSQT) |
| C.e. sibiricus | 24 | 618 | Deer farm in BURQIN county (ALTBRJ) |
| C.e. sibiricus | 28 | 485 | Deer farm in REGIMENT 188 (ALT188) |
| C.e. yarkandensis | 39 | 552 | Deer farm in ALAER (THALR) |
| C.e. yarkandensis | 29 | 311 | Deer farm in XAYA county (THXY) |
| C.e. yarkandensis | 181 | 3089 | Deer farm in KORLA REGIMENT 35 (TH35) |
Microsatellite primers
The seven loci employed were the same as those used by Mahmut et al. (2002), and were synthesized at Sangon Corp. (Shanghai, China). The primer sequences and species source of the seven loci are shown in Table 2. Taq DNA polymerase, deoxynucleotide triphosphate (dNTP), acrylamide and PUC19 DNA markers were purchased from Sangon Corp. Polymerase chain reaction (PCR) amplification was undertaken using a PTC-100TM (Bio-Rad Company, Hercules, CA, US). The electrophoretic apparatus and the electrophoretic trough were types DXY-4 and  , respectively, and made in Beijing by the Liuyi Instrument Factory.
, respectively, and made in Beijing by the Liuyi Instrument Factory.
| Microsatellite loci | Primer sequences (5′→3′) | Chromosome | Annealing temperature | Cycles | Source | References |
|---|---|---|---|---|---|---|
| BM888 | AGGCCATATAGGAGGCAAGCTT | 10 | 49°C | 30 | cattle | Mahmut et al. (2001) |
| CTCGGTCAGCTCAAAACGAG | ||||||
| BM4208 | TCAGTACACTGGCCACCATG | 9 | 56°C | 30 | cattle | Mahmut et al. (2001) |
| CACTGCATGCTTTTCCAAAC | ||||||
| BM5004 | TCTGGAGTGAATGTTTCTGAGG | 20 | 49°C | 30 | cattle | Mahmut et al. (2001) |
| TTGTGATGAGCACCTGAAGG | ||||||
| BM6438 | TTGAGCACAGACACAGACTGG | 1 | 58°C | 28 | cattle | Bishop et al. (1994) |
| ACTGAATGCCTCCTTTGTGC | ||||||
| BM6506 | GCACGTGGTAAAGAGATGGC | 1 | 63°C | 30 | cattle | Bishop et al. (1994) |
| AGCAACTTGAGCATGGCAC | ||||||
| BMS2508 | TTTCTGGGATTACAAAATGCTC | 6 | 55°C | 30 | sheep | Chu et al. (2002) |
| TTTCTTAGGGGAGTGTTGATTC | ||||||
| Celjp0023 | CCACAGGAGACATGGATTCACT | 4 | 54°C | 30 | deer | Slate et al. (2002) |
| CGCAGAACAACTAAGCCAAGTC |
Genomic DNA was extracted from whole blood according to Liu et al. (1997), and examined using 0.8–2% agarose gel. If the DNA band was orderly, clear and only one band, this indicated purity and the band was extracted using hydroxybenzene-chloroform (containing 5% of iso-amylalcohol). PCR was performed in a total volume of 25 µL with the reaction mixture containing 1–5 µL genomic DNA, 2.5 µL 10 × buffer (Mg2+ free), 1.0 µL of MgCl2 (25 mmol/L), 1.5 µL of dNTP (2 mmol/L), 0.6 µL of primers, 20 pmol/µL and 1.5 units of Taq DNA polymerase. The mixture was denatured at 95°C for 5 min followed by 30 cycles at 94°C (denaturation) for 30 s, 49–63°C (annealing) for 45 s and 72°C (extension) for 30 s; the final extension step was performed at 72°C for 10 min.
The PCR products were loaded onto non-denaturing polyacrylamid gel (12%), and 180–200 V were applied for 4–5 h and Sanguinetti silver staining (Shi & Guo 1998) applied to display PCR products of different sizes. Images were scanned and analyzed using Kodak ID Electrophoresis Documentation and Analysis System 120. The size of allele segments and genotypes were determined.
The allele frequency, polymorphism information content (PIC), heterozygosity (H), effective numbers of alleles (E) and rate of homozygosity of alleles (Rh) for each microsatellite loci were assessed for each of the three red deer subspecies as follows.
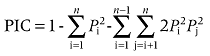
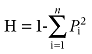

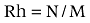
where Pi and pj are the frequencies of allele numbers i and j, respectively, n is the number of alleles at each microsatellite loci, N is the number of animals with homozygosity of alleles and M is the total number of animals in the population.
The genetic distances among all the populations were calculated for each subspecies according to Nei (Sudhir et al. 2001). Two dendrograms were constructed based on either genetic distance by the neighbor-joining method (NJ), utilizing molecular evolutionary genetics analysis software PHYLIP (3.6), or a phylogenetic tree was constructed based on allelic frequencies by maximum likelihood (ML), and the bootstrap value in the tree was estimated by the bootstrap test (Felsenstein 1985).
RESULTS
The microsatellite loci MS2508 and Celjp0023 showed no polymorphism in the nine red deer populations included in this study. The other five microsatellite loci had a total of 22 alleles distributed among the populations as follows: TSHM (20), TS104 (21), TS151 (21), TSQT (13), ALTBRJ (20), ALT188 (15), THALR (21), THXY (20) and TH35 (17). Nine common alleles were found in each of the deer populations (Table 3). Allele frequency, Rh, PIC, E and H for the five microsatellite loci in these nine populations are given in Table 4. The genetic distances among the red deer populations are shown in Table 5 and the dendrograms in Figure 1. Using Chi-square fitness test, the five microsatellite loci were analyzed to determine whether they were in Hardy-Weinberg equilibrium. The Chi-square values of BM6438, BM888, BM4208, BM6506 and BM5004 were 1.76 (P > 0.05), 23.60 (P < 0.05), 239.04 (P < 0.05), 20.68 (P < 0.05) and 0.28 (P > 0.05), respectively. The results suggested that BM6438 and BM5004 were in Hardy-Weinberg equilibrium, while BM888, BM4208 and BM6506 were significantly deviated from Hardy-Weinberg equilibrium.
| Locus | Allele (bp) | TSHM | TS104 | TS151 | TSQT | ALTBRJ | ALT188 | TH35 | TH13 | THSY | Total | Average |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BM6506 | 237 | 0.026 | 0.190 | 0.184 | 0 | 0.053 | 0 | 0.068 | 0.080 | 0.065 | 0.665 | 0.074 |
| 218 | 0.184 | 0.285 | 0.245 | 0 | 0.473 | 0.034 | 0.039 | 0 | 0 | 1.262 | 0.140 | |
| 215 | 0.500 | 0.490 | 0.490 | 0 | 0.211 | 0.251 | 0.435 | 0.540 | 0.612 | 3.529 | 0.392 | |
| 200 | 0.290 | 0.026 | 0.061 | 0.600 | 0.105 | 0.464 | 0.007 | 0.100 | 0.065 | 1.717 | 0.191 | |
| 193 | 0 | 0.009 | 0.020 | 0.400 | 0.158 | 0.251 | 0.451 | 0.280 | 0.258 | 1.826 | 0.203 | |
| BM6438 | 297 | 0.159 | 0.192 | 0.059 | 0 | 0.081 | 0 | 0.132 | 0.125 | 0.107 | 0.855 | 0.095 |
| 289 | 0.250 | 0.200 | 0.255 | 0.177 | 0.189 | 0 | 0 | 0 | 0 | 1.071 | 0.119 | |
| 280 | 0.409 | 0.483 | 0.431 | 0.764 | 0.487 | 0.500 | 0.543 | 0.571 | 0.607 | 4.797 | 0.533 | |
| 264 | 0.182 | 0.125 | 0.255 | 0.059 | 0.243 | 0.500 | 0.325 | 0.304 | 0.286 | 2.278 | 0.253 | |
| BM4208 | 185 | 0.298 | 0.253 | 0.118 | 0 | 0 | 0.056 | 0.336 | 0.331 | 0.441 | 1.838 | 0.204 |
| 175 | 0.149 | 0.094 | 0 | 0 | 0.278 | 0.056 | 0.045 | 0.043 | 0.084 | 0.746 | 0.083 | |
| 166 | 0.064 | 0.133 | 0.470 | 0.250 | 0.194 | 0 | 0.080 | 0.084 | 0 | 1.275 | 0.142 | |
| 164 | 0.383 | 0.373 | 0.294 | 0.083 | 0.500 | 0.500 | 0.356 | 0.438 | 0.390 | 3.317 | 0.368 | |
| 162 | 0.106 | 0.147 | 0.118 | 0.667 | 0.028 | 0.388 | 0.183 | 0.104 | 0.085 | 1.825 | 0.203 | |
| BM5004 | 145 | 0.929 | 0.957 | 0.818 | 0.871 | 0.720 | 0.800 | 0.629 | 0.756 | 0.593 | 7.072 | 0.785 |
| 143 | 0.071 | 0.029 | 0.091 | 0.129 | 0.160 | 0.200 | 0.363 | 0.220 | 0.407 | 1.670 | 0.186 | |
| 135 | 0 | 0.014 | 0.091 | 0 | 0.120 | 0 | 0.008 | 0.024 | 0 | 0.258 | 0.029 | |
| BM888 | 238 | 0.042 | 0 | 0.018 | 0 | 0 | 0 | 0.175 | 0.046 | 0.222 | 0.504 | 0.056 |
| 222 | 0.125 | 0.1120 | 0.130 | 0.222 | 0.290 | 0 | 0.029 | 0.123 | 0 | 1.031 | 0.115 | |
| 208 | 0.292 | 0.168 | 0.130 | 0.156 | 0.065 | 0.107 | 0.457 | 0.354 | 0.472 | 2.199 | 0.244 | |
| 194 | 0.458 | 0.360 | 0.407 | 0.622 | 0.548 | 0.393 | 0.063 | 0.108 | 0.028 | 2.988 | 0.332 | |
| 188 | 0.083 | 0.360 | 0.315 | 0 | 0.097 | 0.500 | 0.276 | 0.369 | 0.278 | 2.278 | 0.253 |
| Locus | Genetic indices | Population | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| TSHM | TS104 | TS151 | TSQT | ALTBRJ | ALT188 | TH35 | THALR | THXY | AVERAGE | ||
| BM6506 | H | 0.6316 | 0.6409 | 0.6622 | 0.4800 | 0.7599 | 0.6582 | 0.6115 | 0.6687 | 0.5494 | 0.6292 |
| PIC | 0.5727 | 0.5957 | 0.6276 | 0.3648 | 0.8422 | 0.6041 | 0.6109 | 0.6633 | 0.5457 | 0.6030 | |
| E | 2.7143 | 2.7848 | 2.9605 | 1.9231 | 4.1657 | 2.9254 | 2.5738 | 3.0186 | 2.2194 | 2.8095 | |
| Rh | 0.4583 | 0.5362 | 0.4615 | 0.8125 | 0.4545 | 0.4286 | 0.3059 | 0.3750 | 0.3889 | 0.4691 | |
| BM6438 | H | 0.7118 | 0.6740 | 0.6805 | 0.3806 | 0.6618 | 0.5000 | 0.5819 | 0.5657 | 0.5383 | 0.5883 |
| PIC | 0.6766 | 0.6451 | 0.6317 | 0.3402 | 0.6164 | 0.3750 | 0.5196 | 0.5055 | 0.4781 | 0.5320 | |
| E | 3.4695 | 3.0677 | 3.1300 | 1.6145 | 2.9568 | 2.000 | 2.3915 | 2.3025 | 2.1657 | 2.5665 | |
| Rh | 0.5417 | 0.5970 | 0.5185 | 0.7500 | 0.4000 | 0.4167 | 0.3509 | 0.4839 | 0.4118 | 0.4967 | |
| BM4208 | H | 0.7270 | 0.7684 | 0.7450 | 0.4861 | 0.7376 | 0.5926 | 0.7753 | 0.6905 | 0.6373 | 0.6844 |
| PIC | 0.7184 | 0.7578 | 0.7235 | 0.4791 | 0.7263 | 0.5170 | 0.7659 | 0.6839 | 0.6325 | 0.6672 | |
| E | 3.6633 | 4.3176 | 3.9216 | 1.9459 | 3.8110 | 2.4546 | 4.4495 | 3.2315 | 2.7574 | 3.3947 | |
| Rh | 0.5385 | 0.5135 | 0.5455 | 0.8788 | 0.4545 | 0.5000 | 0.4509 | 0.5152 | 0.4211 | 0.5353 | |
| BM5004 | H | 0.1327 | 0.0829 | 0.3141 | 0.2248 | 0.4416 | 0.3200 | 0.4729 | 0.3795 | 0.4829 | 0.3168 |
| PIC | 0.1239 | 0.0814 | 0.3028 | 0.1995 | 0.4143 | 0.2688 | 0.3686 | 0.3244 | 0.3663 | 0.2722 | |
| E | 1.1529 | 1.0903 | 1.4578 | 1.2899 | 1.7908 | 1.4706 | 1.8971 | 1.6117 | 1.9337 | 1.5217 | |
| Rh | 0.8462 | 0.9701 | 0.7778 | 0.8519 | 0.7500 | 0.9167 | 0.5886 | 0.7500 | 0.5000 | 0.7724 | |
| BM888 | H | 0.6806 | 0.7000 | 0.7010 | 0.5393 | 0.6015 | 0.5842 | 0.6796 | 0.7096 | 0.6497 | 0.6495 |
| PIC | 0.6392 | 0.6584 | 0.6619 | 0.5181 | 0.5926 | 0.5035 | 0.6769 | 0.6997 | 0.6492 | 0.6222 | |
| E | 3.1304 | 3.3337 | 3.3440 | 2.1704 | 2.5091 | 2.4049 | 3.1211 | 3.4434 | 2.8546 | 2.9235 | |
| Rh | 0.4800 | 0.5469 | 0.4074 | 0.8667 | 0.3889 | 0.3571 | 0.3314 | 0.3939 | 0.4444 | 0.4685 | |
| Mean heterozygosity for all sites | 0.5767 | 0.5732 | 0.6206 | 0.4222 | 0.6405 | 0.5310 | 0.6242 | 0.6028 | 0.5715 | 0.5736 | |
| Mean PIC for all sites | 0.5462 | 0.5477 | 0.5895 | 0.3803 | 0.6384 | 0.4537 | 0.5884 | 0.5754 | 0.5344 | 0.5393 | |
| Effective number of alleles | 2.8261 | 2.9188 | 2.9628 | 1.7888 | 3.0467 | 2.2511 | 2.8866 | 2.7215 | 2.3862 | 2.6432 | |
| Mean rate of homozygotes | 0.5729 | 0.6327 | 0.5421 | 0.8320 | 0.4896 | 0.5238 | 0.4055 | 0.5036 | 0.4332 | 0.5484 | |
| TSHM | TS104 | TS151 | ALTBRJ | ALT188 | TH35 | TH13 | THSY | |
|---|---|---|---|---|---|---|---|---|
| TSQT | 0.56 | 0.57 | 0.55 | 0.64 | 0.50 | 0.59 | 0.56 | 0.61 |
| TSHM | 0.28 | 0.41 | 0.30 | 0.42 | 0.45 | 0.35 | 0.43 | |
| TS104 | 0.34 | 0.28 | 0.44 | 0.44 | 0.28 | 0.38 | ||
| TS151 | 0.28 | 0.47 | 0.43 | 0.39 | 0.47 | |||
| ALTBRJ | 0.44 | 0.49 | 0.47 | 0.52 | ||||
| ALT188 | 0.46 | 0.36 | 0.40 | |||||
| TH35 | 0.17 | 0.19 | ||||||
| TH13 | 0.19 |
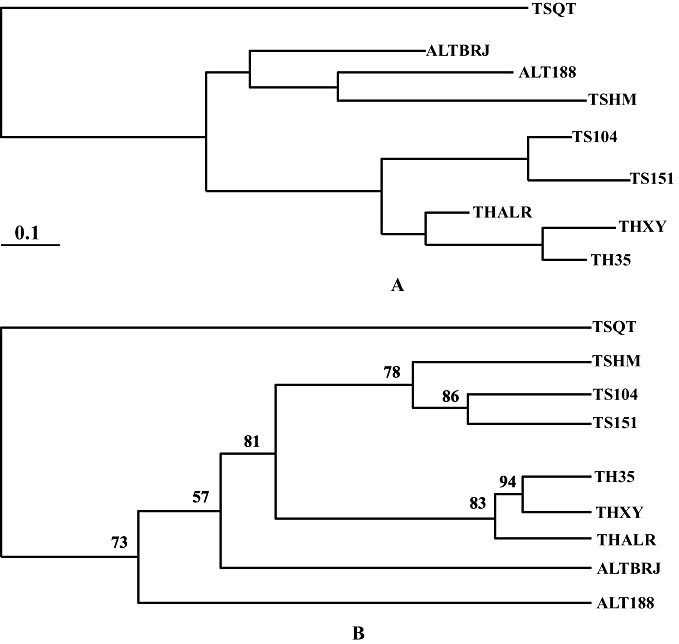
Dendrograms based on genetic distance and allele frequency of nine red deer populations (A: NJ tree; B: ML tree). C.e. songaricus (TSHM, TS104, TS151, TSQT), C.e. sibiricus (ALTBRJ, ALT188), C.e. yarkandensis (THALR, THXY, TH35).
DISCUSSION
Analysis of population genetic background
Deer farms in Korla Regiment 35 began capturing and buying the local wild C.e. yarkandensis from 1959 to 1981. After 1983, as the number of captured wild red deer decreased, the red deer farm bred through self-breeding, and the introduction of a little exogenous red deer. Then the breeding rate was 53.1%. In 1996, a deer farm was established and the breeding rate was 88.3%. In order to improve the quantity and quality of deer product, the red deer of this farm were hybridized with C.e. songaricus and C.e. xanthopygu. Now the red deer number is 1089. The deer farm in ALAER bred C.e. yarkandensis, and the number of captured deer was 76. Thirty-six C.e. yarkandensis from Korla Regiment 35 were introduced to established the ALR deer farm. After that, they were hybridized with C.e. songaricus and C.e. xanthopygu. Now the red deer number is 552. Through capturing and purchase of the local wild C.e. songaricus from Tianshan forest in Qitai, a deer farm was established in 1996 with 128 deer. Mainly through self-breeding, the red deer now number 257. The heterozygosis rate of this population was the lowest in these nine populations. The deer farm in Regiment 151 was established in 1992 with 160 local wild C.e. songaricus, hybridized with C.e. sibiricus from Burqin county. The deer farm in Regiment 104 had C.e. songaricus, established in 1989 with 44 local wild C.e. songaricus; after that hybridization was begun with local wild C.e. songaricus. Now the red deer number is 655. Hong Shan Deer farm in HaMi was established in 1981 with 68 local wild C.e. songaricus; after that they were hybridized with local wild C.e. songaricus. Now the red deer number is 812. In Xaya county and Regiment 188, deer farms were established in 1986 and 1993, respectively. Both deer farms had C.e.sibiricus, about 50 deer. They mainly hybridized with C.e. songaricus. In Xaya county, the red deer in this experiment were bred at different farms.
In this paper we analyzed the genetic information of 418 red deer from the molecular level, including three red deer subspecies of nine populations in XinJiang province. When these farms were established, the deer were captured or purchased. Because the number of red deer was large, there should not have been a bottle neck effect. Following establishment of the deer farms, the founders chose to hybridize with C.e. songaricus to increase body size and hence more meat was produced. However, no hybridized offspring were kept.
Analysis of population genetic diversity in red deer
Polymorphism information content is a good index to weigh polymorphism of microsatellite loci in red deer. Four of seven microsatellite loci were highly polymorphic, but BMS2508 and Celjp0023 showed no polymorphism and BM5004 is a neutral polymorphism (Botstein et al. 1980). The four microsatellite loci with high PIC values are effective markers for the analysis of genetic diversity and phylogenetic relationship among three red deer subspecies (Barker 1994).
Rate of homozygosity of alleles is the proportion of individual homozygosity at the microsatellite loci examined. Inbreeding increases the level of homozygosity for any given gene, so the mean rate of gene homozygosity represents the level of inbreeding in one deer population. In Table 5, the mean rate of gene homozygosity in C.e. songaricus (Rh = 0.6449) was higher than that in Altai red deer (Rh = 0.5067) and in C.e. yarkandensis (Rh = 0.4474). The mean rate of gene homozygosity in Qitai wild red deer population (Rh = 0.8320) was the highest in C.e. songaricus subspecies. The mean rate of gene homozygosity in Regiment 35 red deer population (Rh = 0.4055) was the lowest in C.e. yarkandensis. The overall mean Rh was 0.5484. The Rh in Qitai wild red deer population was very high. There may be a close relation with population size, inbreeding level and consanguinuity. The results are consistent with poor genetic diversity as this population was closed during the long-term.
Genetic heterozygosity (H), reflects genetic variation on some gene loci in each population. Our data indicate that mean H of red deer in Tarim (0.5995) was relatively higher than that in Altay (0.5858) and in Tianshan (0.5482), but the differences are small. Mean H of nine red deer populations was 0.5736. The mean H of red deer in Regiment 35 was the highest (H = 0.6242), which indicates that there are large-scale genetic variations in this population. The results are consistent with the presence of C.e. songaricus in breeding. The research results are close to the report by Polziehn et al. (2000) that mean H is 0.2575–0.5285 utilizing 12 microsatellite loci markers in 11 red deer populations in North America, but higher than the report by Xing et al. (2002) in which polymorphisms of blood proteins were analyzed. Our result is not identical to the report by Mahmut et al. (2001) in which genetic diversity was analyzed utilizing three microsatellite loci markers in Xaya, Lopnur and Qarqan populations of red deer; the results of that report showed that the whole mean H was 0.08 ± 0.02, but mean H was 0.08 ± 0.04 in the Xaya red deer population. This research showed that genetic variation of red deer in Tarim was greater than that in Tianshan and Altay; the genetic diversity of red deer is therefore very rich and large, with potential for genetic breeding in Tarim.
Analysis of phylogenetic relationships in three red deer subspecies
Nei genetic distance (Fig. 1) was calculated among the whole populations according to allelic frequencies. Using the ML tree showed that C.e. songaricus populations in Regiments 104, 151 and Hami were clustered together, noted as cluster A. C.e. yarkandensis populations in Regiment 35, Xaya and Alaer were clustered together, noted as cluster B. Clusters A and B were clustered together, noted as cluster C. Lastly, C.e. sibiricus populations in Burqin, Regiment 188 and cluster C were clustered together. According to the genetic distance and clustering results, genetic distance is very close between C.e. yarkandensis and C.e. songaricus, because they cluster at first in Figure 1, but are not identical to the report by Mahmut et al. (2001). XinJiang red deer are domesticated from wild red deer. The genetic distance is the closest between red deer in Regiment 188 and in Qitai as an outgroup; this is consistent, in that geographic distance is short and activity regions are very close between Regiment 188 and Qitai. This paper showed that every population in three red deer subspecies is clustered in every subspecies, this result is in accordance to origin, history of breeding and distribution by geographic region.




