Effect of liquid whey feeding on fecal microbiota of mature and growing pigs
ABSTRACT
The effect of liquid whey feeding on fecal bacteria and their metabolites was assessed in five pregnant sows and 66 growing pigs. Sows were fed a control diet for 4 weeks (control period) followed by the same diet but with whey feeding (5 L/day/pig) for 4 weeks (whey period). One group of growing pigs was given 267 L of whey per pig (whey group), while the other group was not (control group). In both cases, liquid whey was given separately from control diet. Sows in the whey period had feces showing lower pH, lower ammonia concentration, and larger population sizes of total bacteria, lactobacilli and bifidobacteria. The bacterial gene library analysis indicated that Mitsuokella and Megasphaera were more frequently detected, while Clostridium disporicum were detected less frequently in the whey period. Feces from whey-fed growing pigs showed lower pH than that from control pigs in the early stage of growing. Also, larger populations of total bacteria, lactobacilli and bifidobacteria were recorded in the whey group. From the bacterial gene library analysis, the detection frequency of Lactobacillus reuteri tended to be higher in the whey group. These results indicate that whey feeding influences the hindgut microbiota of pigs, possibly leading to a fermentation shift that is favorable for animal health.
INTRODUCTION
Liquid whey is an attractive feed resource for domestic animals, especially pigs, because liquid feeding is being introduced worldwide with the advantage that feed ingredients are given directly without additional costly processes such as drying. Liquid whey could be considered as a prebiotic feed as well as a protein source, because it contains 5% lactose, which is utilized by beneficial intestinal bacteria such as lactobacilli and bifidobacteria (Coppa et al. 2006). Growth promotion of these lactate-producing bacteria in the pig intestine could improve intestinal health through lactate production and the resultant reduction of intestinal pH, which indirectly prevents the growth of detrimental bacteria, including pathogenic Salmonella and Escherichia coli (Wells et al. 2005). Also, the intestinal immune system could be promoted by selective proliferation of beneficial bacteria (Mitsuoka 2002). Therefore, evaluation of the potency of liquid whey as a prebiotic feed from the viewpoint of intestinal microbiology could be important. In addition, because liquid whey contains lactic acid bacteria that are blended as cheese starters by manufacturers, these bacteria also need to be assessed as possible probiotics for pigs. Furthermore, liquid whey that is rich in proteins, vitamins and minerals is considered to be a nutritionally favorable material as a feed ingredient (Leibbrandt & Benevenga 1991).
Although liquid whey feeding is popular on swine farms and contributes to dairy byproduct consumption all over the world, its merits have not been fully scientifically evaluated. The usefulness of liquid whey feeding in terms of the growth promotion of pigs was initially indicated by Maswaure and Mandisodza (1995). We have also reported the improved growth performance of pigs as the result of liquid whey feeding on a commercial farm and suggested the importance of clarifying some factors involved in this improvement (Kobashi et al. 2009). Intestinal microbiota is a primary factor in modulating pig nutrition and health. Therefore, the aim of the present study was to determine how liquid whey feeding influences the intestinal microbiota of pigs.
MATERIALS AND METHODS
Animals, feeding and sampling
All the experimental pigs were kept at a commercial farm (Inter Farm Co. Ltd, Abashiri, Japan) according to the farm's manual for animal management. Samplings were carried out, following the Act on Welfare and Management of Animals (2005) and Guidelines for Animal Experiments, Hokkaido University (2007).
Five sows (Landrace x Large white x Duroc) in the seventh week of pregnancy were individually housed in stalls and fed an antibiotic-free commercial formula feed with free access to water. The feed (Pigfighter 73; Kumiai Feed Manufacturing, Ohta, Japan) consisted of 62% cereals, 17% oil meals, 14% bran and 7% other ingredients and contained 15% crude protein (CP) and 73% total digestible nutrients (TDN). Sows were assigned to the formula feed for 4 weeks (control period) followed by the same diet but with additional feeding of liquid whey at the level of 5 L/day/pig for 4 weeks (whey period). Liquid whey was fed separately from the formula feed, using a wet feeder. Fresh feces samples were taken from the rectum by grab sampling at the end of the control period and at the 14th and 28th days of the whey period. Feces samples were immediately frozen at −30°C, shipped to the laboratory, and stored at −80°C until analysis.
Sixty-six piglets (LWD) from nine litters were used. The animals, their feeding, and their management were the same as described by Kobashi et al. (2009). In brief, the animals were weaned at 28 days of age (7.8 ± 1.3 kg in body weight), divided into two groups to erase hereditary and sex interference, and given a commercial formula feed (see below) with and without additional supply of liquid whey during the period from 29 to 173 days of age (whey group vs. control group). Whey was given separately from the formula feed as described for sows, but its supply was gradually increased as pigs grew, that is 0.79, 1.5 and 2.0 L/day/pig were fed during the periods of 29–65, 66–104 and 105–173 days of age, respectively. The total supply of liquid whey in the whey group was 267 L/pig. The formula feed for pigs was the starter A (Uruoi Pro; 22.5% CP and 89% TDN), followed by the starter B (Manpuku IF; 18.5% CP and 81.0% TDN), the grower diet (Grower Phase; 16% CP and 79.5% TDN) and then the finisher diet (Finisher Wonder Rich 78IF; 12.5% CP and 78.0% TDN). All the formula feeds were purchased from Nippon Formula Feed, Tomakomai, Japan. Morantel citrate, colistin sulphate, and avilamycin were blended into the starter feed that was given to all pigs until 65 days of age, while no such antibiotics were offered thereafter. At 43, 65, 104 and 139 days of age, fresh feces samples from six pigs randomly selected from each group were taken and stored as described above for sows. This was due to difficulty in continuous sampling from specific individuals over the experimental period.
The experimental whey just before being given to the animals was sampled at the time for fecal sampling (twice in whey period for sows and four times for growing pigs) and subjected to microbial analysis.
MICROBIAL ANALYSIS
DNA extraction
Bacterial DNA was extracted from feces and whey samples by the repeated bead beating plus column (RBB+C) method (Yu & Morrison 2004). In brief, thawed feces was weighed and placed in a sterilized 2 mL screw-capped tube containing 0.4 g of glass beads (diameter: 100–500 µm; Sigma Chemicals, St. Louis, MO, USA). The feces (0.25 g) was mixed with 1.0 mL of lysis buffer (500 mmol/L NaCl, 50 mmol/L Tris-HCl [pH 8.0], 50 mmol/L ethylenediaminetetraacetic acid, and 4% sodium dodecyl sulfate). DNA was extracted by shaking the tube horizontally. The extracted DNA was purified by QIAamp columns (from the QIAamp DNA Stool Mini Kit; Qiagen, Hilden, Germany). The DNA was spectrophotometrically quantified by a plate reader (ARVO MX/Light; PerkinElmer, Waltham, MA, USA).
Real-time polymerase chain reaction
For polymerase chain reaction (PCR) quantification of lactobacilli, the genus-specific primers Lac1f (5′-AGC AGT AGG GAA TCT TCC A-3′) (Walter et al. 2001) and Lab677r (5′-CAC CGC TAC ACA TGG AG-3′) (Heilig et al. 2002) were used. For bifidobacteria, the genus-specific primers Bif164f (5′-GGG TGG TAA TGC CGG ATG-3′) and Bif662r (5′-CCA CCG TTA CAC CGG GAA-3′) (Kok et al. 1996) were employed. Standards for real-time PCR were constructed by amplifying the targeted regions of 16S rDNA of Lactobacillus acidophilus CH2 and Bifidobacterium breve ATCC15700, followed by cloning with E. coli. Both assays were validated for accuracy and reproducibility as described in Koike et al. (2007). Total bacteria were quantified according to the method of Koike et al. (2007).
Denaturing gradient gel electrophoresis
DNA (500 ng) isolated from feces of sows was used as a template to amplify the V3 region of bacterial 16S rDNA by PCR with primers 2 and 3 as described by Muyzer et al. (1993). In brief, touchdown PCR was performed using the rTaq DNA polymerase system (TOYOBO, Osaka, Japan) and the PCR conditions described by Muyzer et al. (1993). The integrity of the PCR products was visually checked by electrophoresis on a 2% agarose gel. The PCR products and the denaturing gradient gel electrophoresis (DGGE) marker (NipponGene, Tokyo, Japan) were separated by the Decode Universal Mutation Detection System (Bio-Rad Laboratories, Hercules, CA, USA) using a 6% (v/v) polyacrylamide DGGE gel with a 30% to 70% gradient of denaturant. Electrophoresis was performed at 90 V for 16 h in Tris-acetate-EDTA buffer (pH 7.4) at a controlled temperature of 60°C. The gel was stained with SYBR green (Takara Bio, Ohtsu, Japan) or silver nitrate and the DGGE gel image was scanned using a LumiVision PRO 400EX image analyzer (Aisin Seiki Co., Ltd, Kariya, Japan). The DGGE band profiles obtained were analyzed by clustering via the unweighted pair group method with mathematical averages (UPGMA) using BioNumerics software (Applied Maths, Inc., Austin, TX, USA).
Bacterial 16S rDNA library
Primers 27F (5′-AGA GTT TGA TCM TGG CTC AG-3′) and 1525R (5′-AAG GAG GTG WTC CAR CC −3′) were used for amplifying bacterial 16S rDNA to construct its library. Templates used for library construction were the mixture of DNA from whey samples, feces of five sows at the end of control and whey periods, and also feces of six growing pigs from control and whey groups at 43 days of age. PCR conditions were as follows: initial denaturation at 94°C for 5 min, then 20 amplification cycles of denaturation at 94°C for 0.5 min, annealing at 58°C for 0.5 min, and extension at 72°C for 1.5 min. A final extension was performed at 72°C for 7 min. The PCR product was purified by agarose electrophoresis and isolated using a QIAEX II Gel Extract Kit (Qiagen, Hilden, Germany). The purified product was then ligated with pGEM T Easy Vector (Promega, San Louis Obispo, CA, USA) and introduced into E. coli JM109. White colonies developed on Luria-Bertani plates with ampicillin, isopropyl beta-D-1-thiogalactopyranoside and X-gal were employed for plasmid isolation using a QIAprep Spin Miniprep Kit (Qiagen) for the following sequencing analysis (Dragon Genomics Center, Takara Bio Inc., Yokkaichi, Japan).
The sequences were compared to those available in the GenBank database using the DDBJ BLAST program (http://www.ddbj.nig.ac.jp/Welcome-e.html). The 16S rDNA clone libraries from the control and whey periods (groups) were compared using the LIBSHUFF computer program (Singleton et al. 2001). The DNADIST program of PHYLIP using the Juke-Cantor model (Felsenstein 1989) was used to generate the distance matrix analyzed by LIBSHUFF. Sequence identity of 97% was used as a criterion to define whether each sequence belonged to a certain species. The sequences (c. 1500 bp) obtained were deposited in the DDBJ nucleotide sequence database under the accession numbers AB506124 through AB506454.
Chemical analysis
Each thawed feces sample (0.1 g) was dispersed into sterilized saline (0.5 mL). After measuring the pH value with an electrode (Horiba, Kyoto, Japan), this solution was centrifuged at 1000 × g for 5 min. The supernatant was used for analyzing short-chain fatty acids (SCFA) by gas-liquid chromatography (Suto 1973), lactate (L- and D-lactate) with a lactic acid assay kit (Megazyme, Wicklow, Ireland), and ammonia nitrogen by a phenol-hypochlorite method (Weatherburn 1967).
Statistical analysis
The data on bacterial abundance and chemical parameters of feces from sows were subjected to one-way analysis of variance by SPSS software (Version 16.0 J, SPSS Inc., Tokyo, Japan), where period was a fixed effect. When significance was detected, multiple comparisons were made by Bonferroni's method. The data from growing pigs at each time point were compared between control and whey groups by Student's t-test. Statistical significance was set at P ≤ 0.05.
RESULTS AND DISCUSSION
Liquid whey as a tested feed
The experimental whey just before being given to pigs showed variations in pH (3.7–4.9) and bacterial abundance (lactobacilli, ×107–8 16S rDNA copies/mL; bifidobacteria, ×105–6 16S rDNA copies/mL) due to season and storage duration. Table 1 shows the bacteria detected in the 16S rDNA mini-library. They belonged to Lactobacillales (50%), Proteobacteria (36%), and Rhodospirillales (14%). Nineteen sequences showing more than 97% identity were identified as those of eight known species. Of these known bacteria, some species of lactate-producing bacteria such as Lactobacillus helveticus, Streptococcus thermophilus, and Lactococcus lactis were considered as starter cultures blended by a cheese manufacturer. Other bacteria were considered as later contaminants during transportation and/or storage. Because all of these species were not detected from the feces of either mature or growing pigs in the present study (see below), they may be regarded as transient microorganisms in the gastro-intestinal tract of the experimental pigs. However, there might be a possibility that these organisms act as probiotics to stimulate indigenous bacteria without notable proliferation of themselves (Ohashi et al. 2007). The possibility is to be further described below.
| Species | No. identified | (%) | Accession no. |
|---|---|---|---|
| Streptococcus thermophilus | 1 | (4.8)‡ | AB506443 |
| Lactobacillus helveticus | 4 | (19.0) | AB506440, AB506444, AB506448, AB506452 |
| Lactococcus piscium | 3 | (14.3) | AB506435, AB506438, AB506451 |
| Lactococcus lactis | 2 | (9.5) | AB506446, AB506447 |
| Rahnella aquatilis | 5 | (23.8) | AB506439, AB506442, AB506445, AB506449, AB506454 |
| Pantoea agglomerans | 1 | (4.8) | AB506441 |
| Acetobacter fabarum | 2 | (9.5) | AB506434, AB506436 |
| Acetobacter orientalis | 1 | (4.8) | AB506450 |
| Total identified | 19 | (90.5) | |
| Total unidentified | 2 | (9.5) | AB506437, AB506453 |
- † Shared sequence identity at 97% or higher level with known bacteria.
- ‡ ‡Numbers of parenthesis are % of total detected.
Effects on mature pigs
Floral changes in sows' feces revealed by PCR/DGGE analysis are shown in Figure 1. Clustering of banding patterns divided fecal community structures into three groups: those that were from sows in the control period and those from the second and fourth weeks of the whey period. In particular, samples clustered together from the fourth week of the whey period were located apart from the other samples, suggesting that liquid whey feeding gradually alters fecal microbiota and significant alteration occurs within 4 weeks of whey feeding.
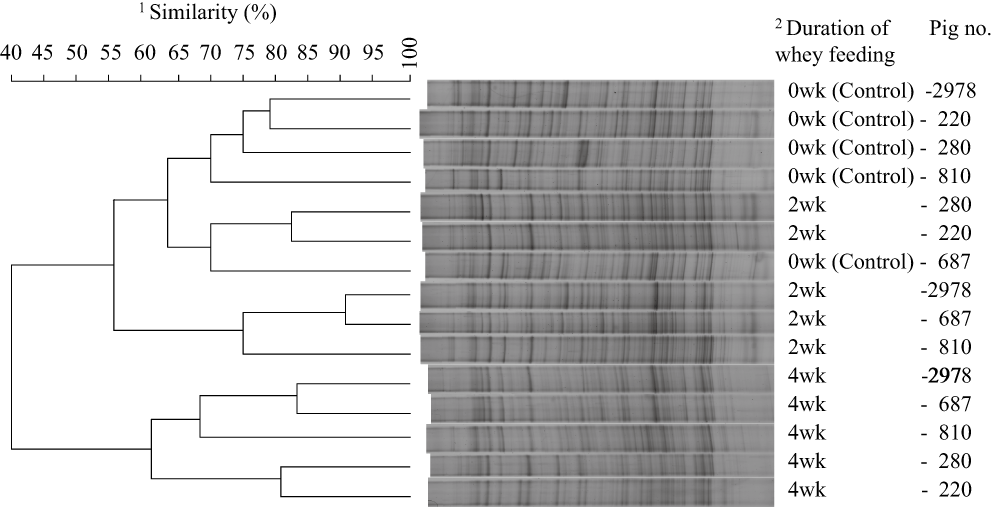
Fecal bacterial community determined by denatured gradient gel electrophoresis and its similarity between sows with and without liquid whey feeding. 1Similarity (%) = 2NAB/(NA+NB). NA and NB are the number of bands detected in sample A and B, respectively, while NAB is the number of bands detected both in sample A and B. 20wk (Control), 2wk and 4wk are samples taken prior to, 2 weeks and 4 weeks after, whey feeding, respectively.
This floral alteration was again evidenced by the analysis of bacterial 16S rDNA libraries as shown in Table 2. The bacterial community retrieved by the library analysis was significantly different between the control and whey periods, shown by the LIBSHUFF test (P < 0.002). Although Firmicutes occupied both libraries, their constituents were different. While the most dominant constituent in the control period was clostridial cluster IV (26.4%), it was replaced by cluster IX (29.5%) in the whey period.
| Phyla/groups | Control† | Whey† | ||
|---|---|---|---|---|
| Firmicutes | ||||
| Clostridiales cluster | ||||
| I | 13 | (14.3)‡ | 7 | (9.0) |
| III | 1 | (1.1) | 0 | (0) |
| IV | 24 | (26.4) | 16 | (20.5) |
| IX | 9 | (9.9) | 23 | (29.5) |
| XI | 8 | (8.8) | 7 | (9.0) |
| XIVa | 17 | (18.7) | 13 | (16.7) |
| Lactobacillales | 14 | (15.4) | 5 | (6.4) |
| Proteobacteria | 4 | (4.4) | 0 | (0) |
| Others | 1 | (1.1) | 7 | (9.0) |
| Total | 91 | (100) | 78 | (100) |
| P value§ | 0.001 | 0.002 | ||
- † Prepared from feces taken prior to and 4 weeks after whey feeding.
- ‡ ‡Numbers in parenthesis are % of total identified.
- § §Represents statistical significance in differences between two libraries.
Although the numbers of known bacterial species (sharing more than 97% 16S rDNA sequence identity) were 24 in the control period and 21 in the whey period, the detection frequency of individual species was different between the control and whey periods (Table 3). For example, no sequence belonging to Mitsuokella or Megasphaera of cluster IX was detected in the control period, but Mitsuokella was detected at 7.5% frequency and Megasphaera was detected at 3.0% frequency in the whey period. Mitsuokella jalaludinii has been considered as a useful bacterium for producing lactate and degrading phytate in the porcine intestine (Lan et al. 2002). Megasphaera elsedenii, a butyrate producer, is also useful for promoting intestinal health (Hashizume et al. 2003), because butyrate stimulates epithelial proliferation in pigs (Cummings et al. 2004). Meanwhile, Clostridium disporicum belonging to cluster I was decreased from 1.2% to nil with liquid whey feeding, suggesting the usefulness of liquid whey to expel this opportunistic bacterium (Woo et al. 2005) from intestinal tracts. However, these bacteria need to be examined in more quantitative analyses.
| Species | Control | Whey | Accession no. | ||
|---|---|---|---|---|---|
| Clostridium sardiniense | 3 | (3.7)‡ | 1 | (1.5) | AB506285, AB506293, AB506322, AB506425 |
| Clostridium butyricum | 2 | (2.4) | 2 | (3.0) | AB506292, AB506318, AB506380, AB506433 |
| Clostridium beijerinckii | 1 | (1.2) | 0 | (0) | AB506291 |
| Clostridium puniceum | 0 | (0) | 1 | (1.5) | AB506414 |
| Ruminococcus flavefaciens | 1 | (1.2) | 0 | (0) | AB506290 |
| Mitsuokella jalaludinii | 0 | (0) | 5 | (7.5) | AB506375, AB506376, AB506403, AB506405, AB506409, |
| Megasphaera elsdenii | 0 | (0) | 1 | (1.5) | AB506398 |
| Megasphaera hominis | 0 | (0) | 1 | (1.5) | AB506427 |
| Clostridium glycolicum | 4 | (4.9) | 3 | (4.5) | AB506297, AB506307, AB506321, AB506338, AB506377, AB506379, AB506390 |
| Clostridium disporicum | 1 | (1.2) | 0 | (0) | AB506357 |
| Clostridium baratii | 0 | (0) | 1 | (1.5) | AB506374 |
| Coprococcus eutactus | 0 | (0) | 1 | (1.5) | AB506372 |
| Roseburia faecalis | 0 | (0) | 1 | (1.5) | AB506423 |
| Kurthia gibsonii | 1 | (1.2) | 1 | (1.5) | AB506298, AB506406 |
| Lactobacillus ruminis | 2 | (2.4) | 0 | (0) | AB506300, AB506301 |
| Lactobacillus amylovorus | 4 | (4.9) | 3 | (4.5) | AB506320, AB506325, AB506331, AB506343, AB506389, AB506395, AB506401 |
| Bacillus bhargavae | 1 | (1.2) | 0 | (0) | AB506347 |
| Acinetobacter seohaensis | 2 | (2.4) | 0 | (0) | AB506287, AB506310 |
| Acinetobacter calcoaceticus | 1 | (1.2) | 0 | (0) | AB506359 |
| Eshcerichia coli | 1 | (1.2) | 0 | (0) | AB506289 |
| Total identified | 24 | (29.3) | 21 | (31.3) | |
| Total unidentified | 58 | (70.7) | 46 | (68.7) | AB506286, AB506288, AB506294, AB506295, AB506296, AB506299, AB506302–AB506306, AB506308, AB506309, AB506311–AB506317, AB506319, AB506323, AB506324, AB506326–AB506330, AB506332–AB506337, AB506339–AB506342, AB506344-AB506346, AB506348–AB506356, AB506358, AB506360–AB506371, AB506373, AB506378, AB506381–AB506388, AB506391–AB506394, AB506396, AB506399, AB506400, AB506402, AB506404, AB506407–AB506413, AB506415–AB506422, AB506424, AB506426, AB506428–AB506432 |
- † Shared sequence identity at 97% or higher level with known bacteria.
- ‡ ‡Numbers in parenthesis are % of total detected.
Quantitative PCR revealed that liquid whey feeding significantly increased (P < 0.05) the numbers of total bacteria, lactobacilli, and bifidobacteria in 2–4 weeks compared with the control period, even though the increase in the percentages of lactobacilli and bifidobacteria (relative abundance of each group in 16S rDNA copy number) was not significant (Table 4). These results indicate that liquid whey could stimulate the growth of intestinal bacteria and that the stimulation is not absolutely selective for lactic acid bacteria. In fact, the above library analysis does not support the selective growth of lactobacilli and bifidobacteria by liquid whey feeding, because no bifidobacteria were detected and no lactobacilli showed an increase in detection frequency with whey feeding (Table 3). Wells et al. (2005) also pointed out that growth stimulation by lactose-rich skim milk is not highly selective for hindgut bacteria in pigs.
| Microbial and chemical parameters quantified | Whey feeding for | ||
|---|---|---|---|
| 0 week (Control) | 2 weeks | 4 weeks | |
| Total bacteria (log 16S rDNA copy/g feces) | 12.07 ± 0.07 a | 12.32 ± 0.16ab | 12.29 ± 0.17b |
| Total lactobacilli (log 16S rDNA copy/g feces) | 9.22 ± 0.63 a | 9.47 ± 0.23ab | 9.77 ± 0.36b |
| Total bifidobacteria (log 16S rDNA copy/g feces) | 7.95 ± 0.48 a | 8.47 ± 0.27b | 8.60 ± 0.66ab |
| Lactobacilli (relative % of total bacteria) | 0.14 ± 0.40 | 0.14 ± 0.13 | 0.30 ± 0.15 |
| Bifidobacteria (relative % of total bacteria) | 0.01 ± 0.01 | 0.01 ± 0.01 | 0.02 ± 0.06 |
| pH | 7.49 ± 0.36 a | 6.92 ± 0.29b | 7.37 ± 0.20ab |
| Total SCFA (mmol/g feces) | 0.73 ± 0.21 | 0.69 ± 0.23 | 0.62 ± 0.18 |
| Acetate (%) | 63.2 ± 7.4 | 58.9 ± 2.9 | 58.7 ± 2.7 |
| Propionate (%) | 21.8 ± 3.7 | 24.7 ± 1.1 | 23.8 ± 2.1 |
| n-Butyrate (%) | 8.2 ± 3.1 | 9.6 ± 1.0 | 9.6 ± 1.7 |
| Total lactate | 7.45 ± 1.95 a | 5.01 ± 0.75b | 4.48 ± 0.38b |
| L-lactate (mmol/g feces) | 4.21 ± 1.56 | 3.26 ± 0.64 | 2.96 ± 0.69 |
| D-lactate (mmol/g feces) | 3.24 ± 0.76 a | 1.75 ± 0.70b | 1.53 ± 0.60b |
| Ammonia (mgN/g feces) | 11.6 ± 5.0 | 6.9 ± 0.8 | 8.4 ± 1.9 |
- †Feces from 5 sows at 0 (Control), 2 and 4 weeks after starting liquid whey feeding was employed for analysis. a,b Values with different superscripts differ significantly at P < 0.05. SCFA, short-chain fatty acids.
Chemical parameters, shown in Table 4, did not reflect the above-mentioned alteration of microbiota. While fecal pH was significantly reduced with 2 weeks of whey feeding, the total SCFA level was unchanged and the lactate level decreased. The SCFA proportion did not significantly differ among the samples. Ammonia tended to be consistently but not significantly lower in the whey period. Overall, the variations in chemical parameters did not correlate with those of microbial parameters, which made speculation difficult. However, one possible explanation for reduced pH without increase of SCFA and lactate may be the accumulation of succinate. Although succinate is rarely detected in the hindgut of pigs under normal conditions, it would be accumulated if the number of succinate-utilizing bacteria is limited and scccinate-producing bacteria are abundantly present. Bacteria increased with whey feeding in the present study may be dominated by active producers of succinate, including lactobacilli, some of which are considered as causative agents of succinate accumulation in pigs (Tsukahara & Ushida 2002). However, the possibility remains to be proven, because succinate production and the related bacteria were not assessed in the present study. The tendency for ammonia level to be lowered with whey feeding may imply the increased cell proliferation of hindgut bacteria which is partly supported by the higher number of bifidobacteria, lactobacilli and total bacteria in feces.
Effects on growing pigs
Bacterial phyla and groups within a phylum detected in the 16S rDNA library for growing pigs are indicated in Table 5. All sequences fell into Firmicutes and about half belonged to Lactobacillales (43.7% in the control group and 50.6% in the whey group). Two libraries from the control and whey groups were not significantly different by the LIBSHUFF test (P = 0.747 and 0.806).
| Phyla/groups | Control | Whey | ||
|---|---|---|---|---|
| Firmicutes | ||||
| Clostridiales cluster | ||||
| I | 3 | (3.4)† | 2 | (2.3) |
| III | 0 | (0) | 1 | (1.1) |
| IV | 14 | (16.1) | 6 | (6.9) |
| IX | 1 | (1.1) | 2 | (2.3) |
| XI | 3 | (3.4) | 3 | (3.4) |
| XIVa | 28 | (32.2) | 28 | (32.2) |
| Lactobacillales | 38 | (43.7) | 44 | (50.6) |
| Others | 0 | (0) | 1 | (1.1) |
| Total | 87 | (100) | 87 | (100) |
| P-value‡ | 0.747 | 0.806 | ||
- † Numbers in parenthesis are % of total identified.
- ‡ ‡Represents statistical significance of difference between two libraries.
Table 6 shows the number of known bacterial species identified in the bacterial 16S rDNA library. Thirty-eight sequences were identified as known species in the control group and 49 sequences were identified in the whey group. Of these, Lactobacillus reuteri showed a higher detection frequency in whey-fed pigs (6.5% in the control group vs. 16.7% in the whey group). This bacterium is known to improve feed conversion by expelling the parasitic Cryptosporidium parvum from pig intestine (Casas & Dobrogosz 1997). Other beneficial actions of L. reuteri, including the production of reuterin, a bacteriocin, and activation of the immune system (Morita & Masaoka 2005), could promote host animal health. Because improvements in feed conversion and survivability of whey-fed growing pigs in the present study were previously reported (Kobashi et al. 2009), the higher abundance of L. reuteri in whey-fed pigs in the early stage of growing might have contributed to these improvements.
| Species | Control | Whey | Accession no. | ||
|---|---|---|---|---|---|
| Clostridium bartlettii | 1 | (1.3)‡ | 2 | (2.4) | AB506166, AB506207, AB506252 |
| Coprococcus catus | 2 | (2.6) | 1 | (1.2) | AB506150, AB506190, AB506281 |
| Ruminococcus obeum | 1 | (1.3) | 4 | (4.8) | AB506135, AB506208, AB506219, AB506224, AB506258 |
| Roseburia inulinivorans | 1 | (1.3) | 0 | (0) | AB506159 |
| Eubacterium rectale | 1 | (1.3) | 0 | (0) | AB506124 |
| Eubacterium halii | 0 | (0) | 1 | (1.2) | AB506265 |
| Lactobacillus amylovorus | 12 | (15.6) | 12 | (14.3) | AB506133, AB506134, AB506141, AB506152, AB506157, AB506170, AB506173, AB506175, AB506179, AB506180, AB506187, AB506189, AB506203, AB506204, AB506210, AB506212, AB506220, AB506239, AB506240, AB506243, AB506247, AB506254, AB506280, AB506284 |
| Lactobacillus reuteri | 5 | (6.5) | 14 | (16.7) | AB506155, AB506158, AB506160, AB506194, AB506196, AB506214, AB506216, AB506218, AB506228, AB506230, AB506231, AB506233, AB506235, AB506259, AB506263, AB506264, AB506268, AB506273, AB506277 |
| Lactobacillus gasseri | 1 | (1.3) | 0 | (0) | AB506144 |
| Lactobacillus johnsonii | 3 | (3.9) | 3 | (3.6) | AB506143, AB506156, AB506176, AB506221, AB506251, AB506283 |
| Streptococcus alactolyticus | 11 | (14.3) | 12 | (14.3) | AB506125, AB506137, AB506142, AB506145, AB506148, AB506149, AB506163, AB506164, AB506168, AB506183, AB506195, AB506213, AB506223, AB506225, AB506226, AB506234, AB506236, AB506238, AB506241, AB506245, AB506249, AB506257, AB506282, |
| Total identified | 38 | (49.4) | 49 | (58.3) | |
| Total unidentified | 39 | (50.6) | 35 | (41.7) | AB506126 – AB506132, AB506136, AB506138-AB506140, AB506146, AB506147, AB506151, AB506153, AB506154, AB506161, AB506162, AB506165 – AB506167, AB506169, AB506172, AB506174, AB506177, AB506178, AB506181, AB506182, AB506184 – AB506186, AB506188, AB506191 – AB506193, AB506197 – AB506202, AB506205, AB506206, AB506209, AB506211, AB506215, AB506217, AB506222, AB506227, AB506229, AB506232, AB506237, AB506242, AB506244, AB506246, AB506248, AB506250, AB506253, AB506255, AB506256, AB506260,-AB506262, AB506266, AB506267, AB506269 – AB506276, AB506278, AB506279 |
- † Shared sequence identity at 97% or higher level with known bacteria.
- ‡ ‡Numbers in parenthesis are % of total detected.
Table 7 shows the changes in bacterial quantity and chemical parameters in the feces of growing pigs taken at four different time points. Quantitative PCR revealed that whey-fed pigs had higher (P < 0.05) numbers of total bacteria, lactobacilli and bifidobacteria at 43 days of age in comparison with control pigs, even though the relative percentage of each bacterial group was not significantly different between control and whey-fed pigs. Any other changes in the bacterial parameters at other time points were not significant. Fecal pH showed a lower value (P < 0.05) at 43 days in whey-fed pigs without alterations in total SCFA and lactate concentration. The results are not straightforward, because fecal concentrations of organic acid do not necessarily reflect bacterial numbers and activities. The inconsistent association of pH with acid concentration may be again excused by the possibility of succinate accumulation as done for the data of matured pigs, which remains to be proven. No particular variations were observed for SCFA molar proportion and ammonia in relation to liquid whey feeding throughout the experimental period.
| Microbial and chemical parameters quantified | Days of age | |||||||
|---|---|---|---|---|---|---|---|---|
| 43 | 65 | 104 | 139 | |||||
| Control | Whey | Control | Whey | Control | Whey | Control | Whey | |
| Total bacteria (log 16S rDNA copy/g feces) | 10.83 ± 0.34 | 11.74 ± 0.25* | nd | nd | 11.22 ± 0.34 | 11.40 ± 0.28 | 11.51 ± 0.22 | 11.36 ± 0.18 |
| Total lactobacilli (log 16S rDNA copy/g feces) | 10.59 ± 0.26 | 11.52 ± 0.25* | nd | nd | 10.76 ± 0.42 | 11.05 ± 0.36 | 10.47 ± 0.30 | 10.49 ± 0.30 |
| Total bifidobacteria (log 16S rDNA copy/g feces) | 7.96 ± 0.36 | 8.41 ± 0.16* | nd | nd | 9.01 ± 0.56 | 8.99 ± 0.48 | 8.66 ± 0.21 | 9.28 ± 0.42 |
| Lactobacilli (relative % of total bacteria) | 65.4 ± 35.1 | 77.3 ± 59.2 | nd | nd | 38.1 ± 18.3 | 48.5 ± 19.6 | 10.7 ± 5.8 | 14.6 ± 5.6 |
| Bifidobacteria (relative % of total bacteria) | 0.2 ± 0.2 | 0.1 ± 0.1 | nd | nd | 1.4 ± 2.2 | 0.6 ± 0.6 | 0.1 ± 0.1 | 1.6 ± 2.5 |
| pH | 6.41 ± 0.23 | 6.21 ± 0.31* | 6.64 ± 0.21 | 6.72 ± 0.15 | 6.75 ± 0.36 | 6.67 ± 0.33 | 6.72 ± 0.31 | 6.92 ± 0.35 |
| Total SCFA (mmol/g feces) | 0.18 ± 0.05 | 0.16 ± 0.04 | 0.16 ± 0.03 | 0.15 ± 0.04 | 0.16 ± 0.05 | 0.17 ± 0.03 | 0.17 ± 0.04 | 0.18 ± 0.05 |
| Acetate (%) | 56.7 ± 5.2 | 56.0 ± 3.6 | 52.6 ± 5.5 | 53.6 ± 3.5 | 51.0 ± 6.9 | 50.6 ± 5.7 | 53.1 ± 3.8 | 55.2 ± 3.8 |
| Propionate (%) | 25.2 ± 2.6 | 25.9 ± 2.1 | 22.7 ± 1.4 | 23.2 ± 1.2 | 24.0 ± 3.3 | 25.5 ± 1.7 | 24.2 ± 2.3 | 22.7 ± 2.2 |
| n-Butyrate (%) | 10.9 ± 2.3 | 11.8 ± 2.5 | 15.9 ± 3.1 | 14.5 ± 2.2 | 15.2 ± 3.9 | 14.8 ± 4.2 | 12.8 ± 1.8 | 12.3 ± 2.2 |
| Total lactate (mmol/g feces) | 10.47 ± 3.69 | 8.99 ± 2.17 | 5.66 ± 3.99 | 5.39 ± 2.11 | 10.82 ± 3.18 | 8.00 ± 3.76 | 8.52 ± 2.44 | 8.10 ± 2.16 |
| L-lactate (mmol/g feces) | 3.83 ± 1.95 | 2.97 ± 1.31 | 1.90 ± 1.33 | 2.19 ± 0.92 | 4.80 ± 1.84 | 3.08 ± 1.67* | 3.08 ± 1.03 | 2.99 ± 1.33* |
| D-lactate (mmol/g feces) | 6.64 ± 1.90 | 6.02 ± 1.10 | 3.76 ± 3.00 | 3.20 ± 1.72 | 6.02 ± 1.51 | 4.92 ± 2.25 | 5.44 ± 1.44 | 5.11 ± 1.03 |
| Ammonia (mgN/g feces) | 0.77 ± 0.31 | 0.79 ± 0.29 | 0.94 ± 0.27 | 0.77 ± 0.34 | 0.63 ± 0.21 | 0.61 ± 0.19 | 0.65 ± 0.22 | 0.58 ± 0.23 |
- †Feces from six pigs of each group were employed for analysis. *Significantly different from control at P < 0.05. nd, not determined; SCFA, short-chain fatty acids.
The fact that fecal bacteria responded to liquid whey only in the early stage of growth may suggest that the efficacy of liquid whey feeding depends on the age of pigs, their original microbiota, and the amount of whey fed. The relative abundance of lactobacilli was much higher at 43 days of age (71.5%) than at 104 days (43.3%) and 139 days (12.7%), implying that the original microbiota at 43 days of age potentiate the effect of liquid whey feeding. Similarly, it is suggested that prebiotics may have a greater impact on fecal bacterial populations in younger infants than in older infants (Nakamura et al. 2009). The amount of whey fed to growing pigs in the present study varied from 0.79 to 2.0 L/days/pig in accordance with animal growth. This was approximately equivalent to 8% of body weight at 43 days and 3% of body weight at 139 days of age. Therefore, the difference in actual whey intake would have differentiated the efficacy of liquid whey.
The flora of growing pigs was apparently different from that of sows, which was supported by the LIBSHUFF test (P < 0.05). In fact, except for Lactobacillus amylovorus, no species of bacteria was shared between mature sows and growing pigs in the present study. Therefore, it is assumed that liquid whey could promote the growth of specific bacteria that varied between growing pigs (e.g. L. reuteri) and sows (e.g. Mitsuokella and Megasphaera). None of the bacteria detected from the experimental pigs originated from the experimental whey. This is again indicative of a possibility that bacteria included in the whey do not multiply in the intestinal tract of pigs but may exert their actions by interacting with indigenous beneficial bacteria as suggested by Ohashi et al. (2007). They demonstrated an increase of indigenous lactobacilli in the porcine cecum with feeding of a probiotic yoghurt strain of Lactobacillus without detecting the strain itself. Therefore, it is not definitive at present whether liquid whey can be a probiotic, or a prebiotic as a lactose carrier, or just a material for delivering other milk-originating nutrients such as proteins, minerals and vitamins (Leibbrandt & Benevenga 1991; Maswaure & Mandisodza 1995). Although whey can be all these beneficial materials for pigs, it should be clarified how the benefits are associated with each other. Then, analytical approach is to be argued, for example unsuitableness of fecal concentration of SCFA as a predictor of production rate has been mentioned (Sakata et al. 2003).
Conclusion
Liquid whey feeding altered the fecal microbiota of pigs, particularly mature pigs. Lactobacilli, bifidobacteria and other beneficial bacterial species showed higher abundances in whey-fed pigs, while one opportunistic species disappeared. Therefore, whey is considered to be useful for promoting animal health and nutrition. Because all speculations made in the present paper are based on the analysis of a limited number of animals and gene sequences, these are to be evidenced by a more extensive dataset.
ACKNOWLEDGMENTS
The authors appreciate the financial support to the present study from the Northern Advancement Center for Science and Technology (the basic research development project 2007–2) and from the Japan Livestock Industry Association (the use development of cheese whey, 2007).




