HLA class II haplotype DRB1*04–DQB1*0301 contributes to risk of familial generalized vitiligo and early disease onset
Summary
Generalized vitiligo is a common autoimmune disorder characterized by white patches of skin and overlying hair caused by loss of pigment-forming melanocytes from involved areas. Familial clustering of vitiligo is not uncommon, and patients and their relatives are at increased risk for a specific complex of other autoimmune diseases. Compared with sporadic vitiligo, familial vitiligo is characterized by earlier disease onset and greater risk and broader repertoire of autoimmunity, suggesting a stronger genetic component, and perhaps stronger associations with specific alleles. To determine whether the major histocompatibility complex (MHC) contributes to the familial clustering of vitiligo and vitiligo-associated autoimmune/autoinflammatory diseases, we performed case–control and family-based association analyses of HLA class II-DRB1 and -DQB1 alleles and haplotypes in affected probands and their parents from 76 European-American Caucasian families with familial vitiligo. Affected probands showed a significantly increased frequency of DRB1*04–DQB1*0301 and a significantly decreased frequency of DRB1*15–DQB1*0602 compared with a large sample of reference chromosomes. Family-based association analyses confirmed these results. Probands with DRB1*04–DQB1*0301 developed vitiligo an average of 13.32 yr earlier than probands with DRB1*15–DQB1*0602. Overall, our results indicate that specific MHC-linked genetic variation contributes to risk of familial vitiligo, although HLA does not completely explain familial clustering of vitiligo-associated autoimmune/autoinflammatory diseases.
Introduction
Generalized vitiligo is an acquired disorder characterized by patches of depigmentation resulting from progressive loss of melanocytes from affected areas (Hann and Nordlund, 2000). Vitiligo is perhaps the most common pigmentation disorder, with prevalence estimates varying from 0.1% to 2% in different geographic regions and ethnic groups (Majumder, 2000). Vitiligo is thought to have an autoimmune etiology, based partly on the frequent occurrence of other autoimmune diseases in patients and their relatives (Ongenae et al., 2003) and previous studies indicate that vitiligo is a component of a specific autoimmune/autoinflammatory diathesis that includes autoimmune thyroid disease, rheumatoid arthritis, psoriasis, adult-onset insulin-dependent diabetes mellitus, pernicious anemia, Addison's disease and systemic lupus erythematosus (Alkhateeb et al., 2003; LaBerge et al., 2005).
The frequency of vitiligo in patients’ first-degree relatives is approximately 7% (Alkhateeb et al., 2003; Majumder et al., 1993), suggesting the importance of genetic factors in disease pathogenesis. Families with multiple relatives affected with vitiligo (familial vitiligo) are characterized by earlier onset of vitiligo and a greater risk and broader spectrum of associated autoimmunity (LaBerge et al., 2005). Inherited susceptibility to vitiligo appears to involve multiple genes that also influence certain other autoimmune/autoinflammatory diseases, consistent with a polygenic, multifactorial model for the inheritance of autoimmune susceptibility and comorbidity.
Many autoimmune/autoinflammatory diseases are associated with specific alleles of one or more genes of the major histocompatibility complex (MHC) at chromosome 6p21. For some disorders, such as type 1 diabetes and rheumatoid arthritis, the MHC may be the strongest genetic risk factor, accounting for over 30% of the total genetic risk (Gregersen, 2003). By comparison, genome-wide linkage studies of vitiligo have shown that the MHC probably is not a major locus underlying susceptibility to vitiligo in Caucasians (Alkhateeb et al., 2002; Fain et al., 2003; Spritz et al., 2004), although there is weak evidence for linkage in Han Chinese families (Chen et al., 2005). Nevertheless, over 20 association studies have reported increased or decreased frequencies of specific class I, class II, or class III MHC alleles in vitiligo patients compared with controls, although only a few results remain significant after statistical corrections and have been confirmed in multiple populations. These include increased frequencies of HLA-A30 (Orecchia et al., 1992; Zhang et al., 2004), HLA-B13 (Metzker et al., 1980; Retornaz et al., 1976; Zhang et al., 2004), HLA-B27 (Poloy et al., 1991; Zhang et al., 2004), HLA-Cw6 (Orecchia et al., 1992; Zhang et al., 2004), DR4 ( Dunston and Halder, 1990; Foley et al., 1983), and HLA-DQw3 (Dunston and Halder, 1990; Orecchia et al., 1992), and decreased frequency of C4A-Q0 (Orecchia et al., 1992; Poloy et al., 1991).
Several studies have indicated that HLA associations with vitiligo may differ between patients with a family history of vitiligo and/or other autoimmune/autoinflammatory diseases versus sporadic or isolated vitiligo, or between patients with earlier versus later onset of vitiligo. Several very small studies have suggested that vitiligo association with the HLA class II antigens DR4 and/or DQw3 is perhaps restricted to patients with early-onset vitiligo, other autoimmune/autoinflammatory diseases, or a family history of vitiligo or other autoimmunity (Dunston and Halder, 1990; Finco et al., 1991; Schallreuter et al., 1993).
In the present study, we report results of case–control and family-based association analyses of HLA class II-DRB1 and -DQB1 alleles in patients with familial vitiligo. A total of 207 Caucasian individuals, including 76 unrelated vitiligo probands each with one or both parents, were genotyped using moderate to high resolution DNA-based typing techniques, making this the largest study of HLA associations with familial vitiligo reported to date. In addition, the parental genotype data made it possible to perform association studies of specific DR–DQ haplotypes. Our results confirm previously reported associations of familial vitiligo with DRB1*04, and also suggest that the vitiligo association with DRB1*04 is stronger when DRB1*04 is combined on the same haplotype with DQB1*0301 than when DRB1*04 is combined on the same haplotype with DQB1*0302. We also present, for the first time, evidence for a negative association between familial vitiligo and haplotype DRB1*15–DQB1*0602.
Results
Case–control comparisons
A comparison of the frequency distributions for HLA-DRB1 and DQB1 alleles between 152 chromosomes from 76 unrelated vitiligo-affected probands (cases) and 3798 reference chromosomes (Klitz et al., 2003) is shown in Figure 1(A, B). Of the 13 DRB1 alleles, three showed significant frequency differences between cases and controls. The frequency of DRB1*04 was significantly higher in cases compared with controls (30.3% versus 17.5%; adjusted P < 0.003), whereas the frequencies of DRB1*15 and DRB1*11 were significantly lower in cases compared with controls (4.6% versus 15.2%, adjusted P < 0.002, and 2.6% versus 9.0%, adjusted P < 0.04 respectively). Two other DRB1 alleles showed suggestive evidence of frequency differences between cases and controls based on nominal significance levels. These were DRB1*10 (2.6% versus 0.7%; nominal P = 0.03) and DRB1*12 (3.3% versus 1.1%; nominal P = 0.03), both of which were relatively infrequent in the study population.
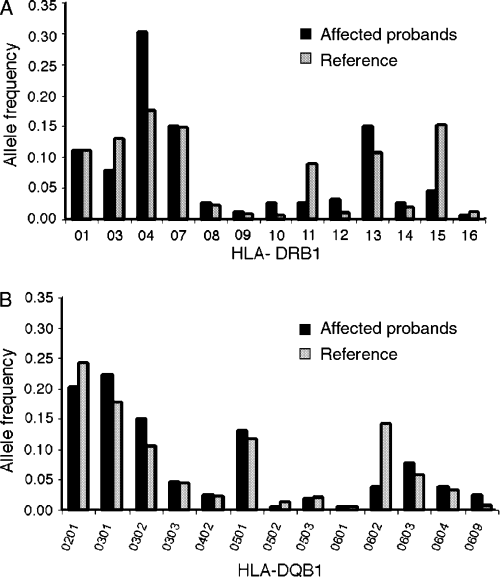
Comparison of the frequencies of (A) HLA-DRB1 and (B) HLA-DQB1 alleles between 152 chromosomes from 76 unrelated vitiligo-affected probands and 3798 reference chromosomes (Klitz et al., 2003). Significance levels were corrected for multiple comparisons.
Of the 13 DQB1 alleles, only DQB1*0602 was significantly decreased in cases compared with controls (4.0% versus 14.4%; adjusted P < 0.0008). None of the DQB1 alleles showed significantly increased frequencies in cases compared with controls after adjusting for multiple comparisons. However, there was suggestive evidence for an increased frequency of a rare DQB1*0609 allele in cases compared with controls (2.6% versus 0.7%; nominal P = 0.03).
Of 169 possible DR–DQ haplotypes, only 22 were observed in vitiligo probands, and only eight were present in above 5% frequency in either or both cases and controls. A comparison of DR–DQ haplotype frequencies between cases and controls is shown in Figure 2 for the eight most common haplotypes. Of the two most common DRB1*04 haplotypes, only DRB1*04–DQB1*0301 showed significantly increased frequency in cases compared with controls (15.1% versus 6.5%; adjusted P < 0.003). The frequency of DRB1*04–DQB1*0302 was also higher among cases, but the difference was not statistically significant.
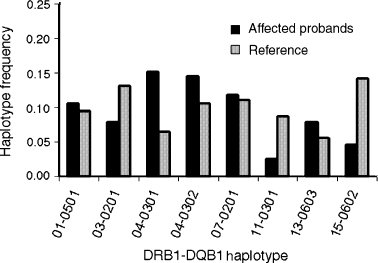
Comparison of the frequencies of eight common DRB1–DQB1 haplotypes between unrelated vitiligo-affected probands and 3798 reference chromosomes. Significance levels were corrected for multiple comparisons.
Consistent with the single-locus analyses, the frequency of the HLA DRB1*15–DQB1*0602 haplotype is considerably lower in cases compared with controls (4.6% versus 14.2%; adjusted P < 0.003). The frequency of DRB1*11–DQB1*0301 is also lower in cases than in controls (2.6% versus 8.7%); however, the difference is of borderline significance (P < 0.06) after correction for multiple comparisons.
Because of strong linkage disequilibrium across the HLA class II region, the DQA1 allele carried by a DRB1–DQB1 haplotype can be predicted with considerable accuracy. Specifically, 98% of DRB1*04–DQB1*0301 haplotypes carry DQA1*0302, 91% of DRB1*04–DQB1*0302 carry DQA1 0301, 99.8% of DRB1*15–DQB1*0602 carry DQA1*0102, and 100% of DRB1*11–DQB1*0301 carry DQA1 0501 (Klitz et al., 2003). Therefore, the most likely high-risk haplotype is DRB1*04–DQA1*0302–DQ B1*0301, and the most likely low-risk haplotype is DRB1*15–DQA1*0102–DQB1*0602.
Subgroup comparisons
The mean age of onset of vitiligo among 76 unrelated probands was 15.93 yr (SD = 10.42). As shown in Table 1, we observed extensive variability in age of onset associated with different DRB1–DQB1 haplotypes. Remarkably, the mean age of onset is considerably earlier in affected probands with the high-risk DRB1*04–DQB1*0301 haplotype (13.85 yr, SD 8.78) compared with affected probands with the low-risk DRB1*15–DQB1*0602 haplotype (27.17 yr, SD = 12.62). This difference is highly significant (P = 0.007).
| DRB1–DQB1 haplotype | Average age of onset of vitiligo (SD) |
|---|---|
| All affected probands | 15.93 (10.42) |
| 01-0501 | 16.36 (6.57) |
| 03-0201 | 16.00 (10.84) |
| 04-0301 | 13.85 (8.78) |
| 04-0302 | 14.83 (10.80) |
| 07-0201 | 18.29 (10.38) |
| 11-0301 | 14.38 (8.94) |
| 13-0603 | 15.40 (10.70) |
| 15-0602 | 27.17 (12.62) |
There were no significant differences between 57 affected probands from families in which the proband or at least one other family member is affected with vitiligo-associated autoimmune/autoinflammatory disease (autoimmune thyroid disease, rheumatoid arthritis, psoriasis, adult-onset type 1 diabetes, pernicious anemia, Addison's disease, and systemic lupus erythematosus; Alkhateeb et al., 2003; LaBerge et al., 2005) and 19 affected probands from families without these autoimmune diseases (Figure 3). However, there was a threefold increase in DRB1*15–DQB1*0602 in vitiligo families without other autoimmunity (7.9% compared with 2.6% in families with autoimmune/autoinflammatory disease; P = 0.165), which may be of interest because of the strong negative association of this haplotype with type 1 diabetes (Redondo et al., 2001).
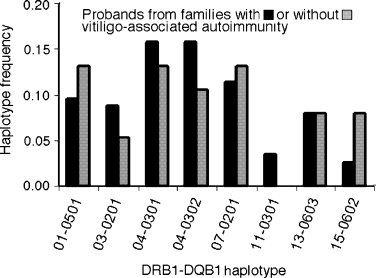
Frequencies of DRB1–DQB1 haplotypes among vitiligo-affected probands from families in which the proband or other family members are affected with vitiligo-associated autoimmune/autoinflammatory diseases (autoimmune thyroid disease, rheumatoid arthritis, psoriasis, adult-onset insulin-dependent diabetes mellitus, pernicious anemia, Addison's disease and systemic lupus erythematosus; Alkhateeb et al., 2003; LaBerge et al., 2005) compared with vitiligo-affected probands from families without vitiligo-associated autoimmune/autoinflammatory diseases.
Family-based association tests
There were no significant differences in allele or haplotype frequencies between the 76 vitiligo-affected probands and 26 vitiligo-affected parents from the same families. When compared with their affected offspring, affected parents showed a similarly increased frequency of DRB1*04–DQB1*0301 (13.5%) and similarly decreased frequency of DRB1*15–DQB1*0602 (5.8%). By comparison, the frequencies of DRB1*04–DQB1*0301 and DRB1*15–DQB1*0602 among AFBAC chromosomes were 7.0% and 11.6%, respectively, similar to what was found among reference chromosomes (6.5% and 14.2%, respectively).
Results of the transmission disequilibrium test of common haplotypes are shown in Figure 4. Overall, there was little or no evidence for deviations from the expected 50% transmission of haplotypes to affected offspring from heterozygous parents. However, there were marked differences between affected and unaffected parents in transmission frequencies for the same haplotype. When the analysis is restricted to unaffected parents, there is significant evidence for increased transmission of DRB1*04–DQB1*0301 (70.43%, P < 0.04) and decreased transmission of DRB1*15–DQB1*0602 (24.37%, P < 0.02), suggestive of recessive inheritance of MHC-linked vitiligo susceptibility.
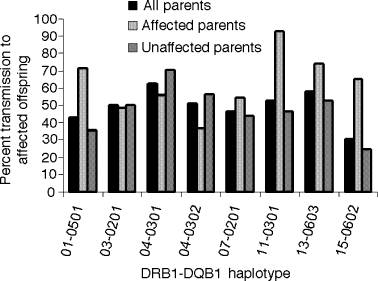
Transmission frequencies of common DRB1–DQB1 haplotypes from affected and unaffected parents to affected offspring.
Discussion
Our findings show that familial vitiligo is positively associated with HLA class II haplotype DRB1*04–DQB1*0301 and negatively associated with haplotype DRB1*15–DQB1*0602. Remarkably, the onset of vitiligo in patients with the high-risk DRB1*04–DQB1*0301 haplotype was approximately 13 yr earlier than in patients with the low-risk DRB1*15–DQB1*0602 haplotype. This is of particular interest in view of our previous finding of an earlier age of disease onset in familial vitiligo when compared with sporadic, unselected vitiligo. The observed association with HLA DRB1*04–DQB1*0301 may also contribute to the greater risk and expanded repertoire of associated autoimmune/autoinflammatory disease in familial vitiligo (Table 2A, B; LaBerge et al., 2005).
| Vitiligo-associated autoimmunity | High risk |
|---|---|
| (A) Sporadic vitiligo patients and their relatives | |
| Autoimmune thyroid disease | |
| Hypothyroidism | Weak |
| Hyperthyroidism | DRB1*03 |
| Pernicious anemia | None reported |
| Addison's disease | DRB1*03 |
| Systemic lupus erythematosus | DRB1*03, C4AQ0 |
| (B) Familial vitiligo but not in sporadic vitiligo | |
| Psoriasis | Cw*0602 |
| Rheumatoid arthritis | DRB1*04 |
| Adult insulin-dependent diabetes | DRB1*03–DQB1*0201 DRB1*04–DQB1*0302 |
- aFor individual disease associations see Peterson et al., 2000; Simmonds and Gough, 2004; Yu and Whitacre, 2004.
- bFor individual disease associations see Pozzilli and Di Mario, 2001; Newton et al., 2004; Rahman and Elder, 2005.
Our results thus confirm reports of vitiligo association with DR4 in white American (Foley et al., 1983), Dutch (Zamani et al., 2001) and Turkish (Tastan et al., 2004) patients, in a subset of German patients with familial vitiligo (Schallreuter et al., 1993), and with DR4 and DQw3 in black Americans (Dunston and Halder, 1990). Furthermore, our results suggest that the association with DRB1*04 differs depending on the DQw3 subtype (DQB1*0301 or DQB1*0302) present on the same haplotype with DRB1*04. Nevertheless, it is important to point out that a number of studies have found no evidence for vitiligo association with HLA DR4; in patients from Hungary (Poloy et al., 1991), northern Italy (Finco et al., 1991; Lorini et al., 1992; Orecchia et al., 1992), Japan (Ando et al., 1993), Amsterdam (Venneker et al., 1993), Oman (Venkataram et al., 1995), Kuwait (al-Fouzan et al., 1995), and Slovakia (Buc et al., 1998). Vitiligo associations have also been reported for several other MHC loci, but the vitiligo-associated alleles at these different loci are not known to be associated with each other or with any ancestral MHC haplotype (Bugawan et al., 2000; Yunis et al., 2003).
Inconsistencies between populations and even between studies of the same population are common in case–control disease association studies, and may be explained by weak genetic effects or complex gene–gene or gene–environmental interactions, population stratification, or genetic differences between populations (Hirschhorn et al., 2002). We addressed some of these problems by combining case–control and family-based study designs and analyses. Our study was restricted to Caucasian families with multiple relatives affected with vitiligo, based on our previous findings suggesting that allelic associations may be stronger in these families (LaBerge et al., 2005). The availability of a large sample of reference chromosomes allowed us to maximize statistical power by performing initial association tests using a case–control approach. The results were then validated using a family-based approach, which is insensitive to artifacts resulting from population stratification.
Differing HLA associations may reflect genetic heterogeneity between different subgroups of patients with vitiligo, combined with differences in inclusion and exclusion criteria in different studies. We and others have shown that unselected, and typically sporadic, vitiligo patients are at increased risk for specific autoimmune/autoinflammatory diseases, including autoimmune thyroid disease, pernicious anemia, Addison's disease, and systemic lupus erythematosus (Alkhateeb et al., 2003; LaBerge et al., 2005). As shown in Table 2A, none of these disorders show strong association with DRB1*04–DQB1*0301, although there have been reports of weak association between DRB1*04 and autoimmune hypothyroidism (Simmonds and Gough, 2004) and between DRB1*0404–DQB1*0302 and Addison's disease (Myhre et al., 2002; Yu et al., 1999). Three of these disorders, autoimmune hyperthyroidism, Addison's disease, and systemic lupus erythematosus, are associated with DRB1*0301, although the frequency of this allele was not increased in the familial vitiligo patients in our study. Results of other studies of the association of vitiligo with DRB1*0301 vary, with the majority of studies reporting no association (Ando et al., 1993; Buc et al., 1998; Foley et al., 1983; Orecchia et al., 1992; Schallreuter et al., 1993; Venkataram et al., 1995; Zamani et al., 2001), a few studies reporting weak negative association (Dunston and Halder, 1990; Finco et al., 1991; Venneker et al., 1993; Zettnig et al., 2003), one study reporting weak positive association (Tastan et al., 2004), and another study reporting weak positive association in vitiligo patients with other autoimmunity (Poloy et al., 1991). The differences in HLA associations between vitiligo versus the specific vitiligo-associated autoimmune/autoinflammatory diseases suggest that variation within the MHC does not contribute significantly to the increased frequencies of these other diseases in unselected vitiligo patients and their relatives.
In families with multiple relatives affected with vitiligo, the repertoire of associated autoimmune/autoinflammatory disease is expanded, including psoriasis, rheumatoid arthritis and adult-onset, insulin-dependent diabetes, in addition to autoimmune thyroid disease, pernicious anemia, Addison's disease and lupus found in unselected vitiligo patients and their relatives (LaBerge et al., 2005). As shown in Table 2B, there is strong and consistent evidence for association of DRB1*04 with rheumatoid arthritis (Newton et al., 2004) and with adult-onset, insulin-dependent (type 1) diabetes (Pozzilli and Di Mario, 2001), although in the latter case the strongest association is with DRB1*04–DQB1*0302, not with DRB1*04–DQB1*0301. Interestingly, the DRB1*15–DQB1*0602 haplotype is associated with dominant protection in type 1 diabetes (Redondo et al., 2001), which could relate to our finding of negative association between DRB1*15–DQB1*0602 and familial vitiligo. The association between psoriasis and HLA-Cw*0602 may also be of interest, based on previous reports of vitiligo association with this allele (Orecchia et al., 1992; Zhang et al., 2004).
Overall, our results indicate that variation within the MHC contributes to the unique characteristics of familial vitiligo, including earlier onset of the vitiligo phenotype and a greater risk and broader spectrum of other autoimmune/autoinflammatory phenotypes compared with non-familial vitiligo. It remains unclear whether and how these findings relate to singleton vitiligo patients, who comprise the majority of cases. We suggest that one or more genes of the MHC may modulate the expression of other autoimmunity susceptibility genes; for example, by influencing tissue specificity and the rate of progression of clinical disease in susceptible individuals. HLA class II genotypes may thus have implications for predicting the occurrence of vitiligo and associated autoimmune/autoinflammatory diseases in patients and their relatives.
Materials and methods
This study was approved by the Colorado Multiple Institutional Review Board (COMIRB) and the South Thames Regional Multicentre Research Ethics Committee (MREC). Written informed consent was obtained from all study participants. Families with two or more relatives affected with vitiligo were ascertained principally from the Vitiligo Society (UK) and the National Vitiligo Society (USA), as described elsewhere (Fain et al., 2003). Phenotypes were checked carefully by history, lesion maps and, in most cases, physical examination and/or photographs; individuals for whom the phenotype was at all questionable were excluded.
DNA was prepared from peripheral blood by standard methods. A total of 207 individuals from 76 European-American Caucasian multiplex vitiligo families were genotyped for HLA-DRB1 and -DQB1, with each family consisting of an affected proband and at least one affected or unaffected parent. For 55 of the families, both parents were genotyped, and for 21 families one parent was genotyped. HLA-DRB1 and -DQB1 genotyping was performed using polymerase chain reaction (PCR)-based, sequence-specific oligonucleotide probe (SSOP) hybridization systems (Dynal Biotech, Inc., Oslo, Norway) as described (Noble et al., 1996). Briefly, oligonucleotide probes specific for polymorphisms in an HLA locus were immobilized in linear array on a nylon membrane. The polymorphic exons 2 of HLA-DRB1 and -DQB1 were amplified by PCR with biotinylated oligonucleotide primers and PCR products were hybridized to the array. Probe binding was visualized using streptavidin–horseradish peroxidase (HRP) and a colorimetric substrate, tetramethylbenzidine (TMB). Binding patterns were translated to genotypes using genotyping software.
HLA allele and haplotype frequencies for 76 unrelated affected probands were compared with published reference standards derived from 1899 European–American bone marrow donors (Klitz et al., 2003) using an exact test. The allele and haplotype frequencies for the reference sample are very similar to the frequencies reported for other smaller samples of healthy individuals of Northern European descent (Carrington et al., 1994; Darke et al., 1998), and also did not differ significantly from 43 non-transmitted chromosomes (affected family-based controls; AFBAC chromosomes) from unaffected parents in the 76 vitiligo families under study. To adjust for multiple comparisons, nominal significance levels were multiplied by the number of common (>0.05 frequency in affected probands) alleles or haplotypes, plus one additional allele/haplotype at each locus to account for the collection of rare alleles (multipliers of 12 and 9, for single locus and 2-locus analyses, respectively). The transmission disequilibrium test (TDT) implemented in unphased (Dudbridge, 2003) software was used for family-based association tests (Clayton, 1999; Cordell and Clayton, 2002; Spielman et al., 1993).
Acknowledgements
We thank the Vitiligo Society (UK) and the National Vitiligo Foundation (USA) for their enthusiastic participation in this study. This work was supported by grants AR45584, A146374, and DK57538 from the National Institutes of Health.