Derivation of a novel undifferentiated human foetal phenotype in serum-free cultures with BMP-2
Abstract
Skeletal stem and progenitor populations provide a platform for cell-based tissue regeneration strategies. Optimized conditions for ex vivo expansion will be critical and use of serum-free culture may allow enhanced modelling of differentiation potential. Maintenance of human foetal femur-derived cells in a chemically defined medium (CDM) with activin A and fibroblast growth factor-2 generated a unique undifferentiated cell population in comparison to basal cultures, with significantly reduced amino acid depletion, appearance and turnover, reduced alkaline phosphatase (ALP) activity and loss of type I and II collagen expression demonstrated by fluorescence immunocytochemistry. Microarray analysis demonstrated up-regulation of CLU, OSR2, POSTN and RABGAP1 and down-regulation of differentiation-associated genes CRYAB, CSRP1, EPAS1, GREM1, MT1X and SRGN as validated by quantitative real-time polymerase chain reaction. Application of osteogenic conditions to CDM cultures demonstrated partial rescue of ALP activity. In contrast, the addition of bone morphogenetic protein-2 (BMP-2) resulted in reduced ALP levels, increased amino acid metabolism and, strikingly, a marked shift to a cobblestone-like cellular morphology, with expression of SOX-2 and SOX-9 but not STRO-1 as shown by immunocytochemistry, and significantly altered expression of metabolic genes (GFPT2, SC4MOL and SQLE), genes involved in morphogenesis (SOX15 and WIF1) and differentiation potential (C1orf19, CHSY-2,DUSP6, HMGCS1 and PPL). These studies demonstrate the use of an intermediary foetal cellular model for differentiation studies in chemically defined conditions and indicate the in vitro reconstruction of the mesenchymal condensation phenotype in the presence of BMP-2, with implications therein for rescue studies, screening assays and skeletal regeneration research.
Introduction
The application of stem/progenitor cells to injury or disease management will require sufficient cell number, thus necessitating derivation of ex vivo expansion protocols as the native stem cell resource is limited by their availability, typically less than one in every 40,000 cells [1, 2]. Expansion of cultures in vitro must be tempered with the ability of the cells to maintain a proliferative capacity and stem/progenitor phenotype prior to targeted lineage differentiation. Most tissue culture techniques utilize foetal calf serum (FCS), a complex undefined mixture of factors, although batch variability has been shown to have significant effects on expansion kinetics of human bone marrow stromal cells (hBMSCs) [3]. The use of human-derived serum has been shown to support hBMSC and chondrocyte cell growth; however, these studies are limited by issues of cost and availability [4, 5]. Use of a serum-free growth medium may permit better modelling of differentiation potential [6–8], and a serum-free chemically defined medium (CDM) developed by Johansson and Wiles [6] was later optimized for undifferentiated human embryonic stem cells (hESC) culture by Vallier and colleagues with the addition of either activin A or Nodal as inhibition of activin/Nodal signalling, though not Nodal alone, resulted in increased differentiation [9]. Following supplementation with fibroblast growth factor-2 (FGF-2), hESCs maintained pluripotency in long-term cultures suggesting FGF-2 as a competence factor in the activin/Nodal pathway [9].
Many tissue regeneration strategies rely on the application of stimulatory agents to induce differentiation following expansion [10, 11] and various factors have been applied to mesenchymal cell populations [12–14]. Ascorbic acid 2-phosphate (ascorbate), acts as an enzyme co-factor for collagen synthesis and has been shown to enhance the osteogenic response of human osteoblasts in vitro[15] as has dexamethasone, a glucocorticoid hormone which promotes the catabolism of carbohydrates, fats and proteins and antagonizes insulin signalling [12, 16–18]. The bone morphogenetic proteins (BMPs) are potent mitogens for the formation of new skeletal tissue, and have been extensively studied for their effects on osteogenic and chondrogenic differentiation [19–21] and the proliferative ability of cells [19, 20, 22–25]. First described as components of demineralized bone matrix by Urist and coworkers [26], BMPs are key in the signal transduction of SMAD proteins 1, 5 and 8 which up-regulate runt-related transcription factor 2 (RUNX2, also referred to as CBFA1 or core-binding factor 1) and result in downstream up-regulation of bone matrix proteins type I collagen, osteocalcin and osteopontin, necessary for osteoblast maturation [27].
Various studies have reported on the osteogenic properties of human foetal bone cells. Harris and coworkers demonstrated increased levels of alkaline phosphatase (ALP) and osteocalcin in an immortalized human foetal osteoblastic cell line in the presence of vitamin D3[28] although studies with primary bone cells from foetuses at 11–14 weeks after conception (WPC) by Campagloni et al.[29] or 13–16 WPC by Montjovent et al.[30], indicated the proliferation of foetal bone cells was increased in the presence of dexamethasone and showed that following treatment with vitamin D3, ascorbic acid and β-glycerolphosphate, foetal populations displayed enriched ALP activity, with up-regulation of type I collagen, ALP, osteocalcin and RUNX2 gene expression [30]. In addition, we have demonstrated an osteoprogenitor phenotype of human foetal femur-derived cells at 7.5–11 WPC with expression of type I collagen, activated leukocyte cell adhesion molecule (ALCAM or CD166), STRO-1, ALP activity and BMP receptor 1A in basal conditions, and up-regulation of non-collagenous bone proteins osteopontin and osteocalcin in the presence of ascorbate and dexamethasone [31].
Human foetal femur-derived cells provide an important intermediary cellular model as a pre-natal but non-embryonic source, between hESC and adult cell populations. Delineation of their osteogenic potential for application to modelling studies will be critical. The current studies, therefore, set out to model the effects of potent osteogenic growth factors ascorbate/dexamethasone and BMP-2 on human foetal femur-derived cells in a serum-free CDM, to provide greater understanding therein for ex vivo expansion, application to growth factor screening and skeletal tissue engineering.
Materials and methods
Reagents were obtained from Sigma-Aldrich (St. Louis, MO, USA) including α-MEM (minimum essential medium, α-modification, M0644), bovine serum albumin (BSA, A1470), monothioglycerol (M6145), PBS (phosphate buffered saline, P4417), trypsin-EDTA (ethylenediamine tetra-acetic acid, T4174), Tris-EDTA (Tris-hydrochloric acid EDTA, T9285), TRITON® X-100 (X100), alkaline buffer solution (A9226), ALP assay kit with Sigma 104® phosphatase substrate (104–0) and AP standard (104–1), IGEPAL® CA-630 (I3021) and primers for real-time quantitative RT-PCR (qRT-PCR). FCS (10106169), Iscove’s modified Dulbecco’s medium (IMDM, 21980–032), F-12 (Ham’s) nutrient mixture (31765–027), Lipid 100× mix (11905–031), TRIzol reagent (15596–018), Super-Script First-strand synthesis system for PCR (11904–018), Cell Tracker Green™ CMFDA (5-chloromethyl-fluorescein diacetate, C7025), ethidium homodimer-1 (E1169) and DAPI (4′,6-diamidino-2-phenylindole, D3571) were purchased from Invitrogen (Paisley, UK). Collagenase B (1088807), insulin (1376497) and transferrin (652202) were procured from Roche (Manheim, Germany). Activin A (120–14) and FGF-2 (100–18B) were purchased from PeproTech EC (London, UK). DNA-free RNA Kit (R1013) was obtained from Zymo Research Corporation (Orange, CA, USA). BMP-2 protein was obtained from Prof. Walter Sebald, University of Würzburg, Germany. Undiluted culture supernatant was derived from the STRO-1 hybridoma (IgM) provided by Dr. J. Beresford, University of Bath. Type I collagen polyclonal rabbit antibody was a gift from Dr. Larry Fisher, of the National Institutes of Health (NIH). Type II collagen polyclonal rabbit antibody (234187) was purchased from Calbiochem (Nottingham, UK). SOX-2 (AB5603) and SOX-9 (AB5535) polyclonal rabbit antibodies were obtained from Chemicon International, Inc. (Temecula, CA, USA). Goat antimouse IgM fluorescein isothiocyanate (FITC)-conjugate (115–096-075) and goat anti-rabbit IgG TRITC (tertramethyl rhodamine isothiocyanate)-conjugate (111–026-047) secondary antibodies were purchased from Jackson ImmunoResearch Labs., Inc. (West Grove, PA, USA and Stratech Scientific Ltd., Soham, Cambridgeshire, UK). Power SYBR® Green PCR Master Mix (4367659) was purchased from Applied Biosystems (Foster City, CA, USA). All images were captured on a Carl Zeiss Axiovert 200 microscope with Axiovision software (version 4.5) via an AxioCam HR digital camera for phase images or via an AxioCam MRc with appropriate filters for fluorescence microscopy (Carl Zeiss Ltd., Welwyn Garden City, UK).
Cell culture
Human foetal tissue was obtained with informed and written consent following termination of pregnancy, according to guidelines issued by the Polkinghorne Report [32] and ethical approval from the Southampton & South West Hampshire Local Research Ethics Committee for use of tissue at 7.5–11 WPC. Tissue was obtained from both sexes (as obtained from placental identification), and foetal age was determined by measuring foot length, with a total of 15 human foetal samples utilized (mean 8.8 ± 0.7 WPC) as shown in Table 1. Foetal cells were isolated following collagenase B digestion of femurs as previously described [31]. Primary cells (passage 0) were established in basal medium (α-MEM containing 10% FCS) at 37°C with 5% CO2.
| Foetal sample | Foot length | Age (WPC) |
|---|---|---|
| H549 | 10.0 | 11.0 |
| H589 | 6.5 | 8.5 |
| H815 | 6.5 | 8.5 |
| H827 | 7.5 | 9.0 |
| H858 | 6.0 | 8.5 |
| H860 | 6.5 | 8.5 |
| H948 | 7.0 | 9.0 |
| H988 | 6.0 | 8.5 |
| H993 | 6.5 | 8.5 |
| H1052 | 5.0 | 7.5 |
| H1054 | 7.0 | 9.0 |
| H1119 | 7.0 | 9.0 |
| H1124 | 7.0 | 9.0 |
| H1126 | 6.5 | 8.5 |
| H1148 | 6.0 | 8.5 |
Chemically defined medium
CDM comprising 50% IMDM/50% F-12, supplemented with 5 mg/ml BSA, Lipid 100× at 1% concentration, 450 μM monothioglycerol, 7 μg/ml insulin and 15 μg/ml transferrin was used with the addition of activin A and FGF-2 where appropriate at 10 ng/ml and 12 ng/ml, respectively [9]. Primary cultures were established as above then seeded to well plates. After 24 hrs, cells were washed twice with PBS then transferred to either CDM with activin A/FGF-2 or back to basal medium conditions with media changes every 48 hrs until harvest at 7 days for metabolic or biochemical analysis.
Osteogenic modulatory factors with CDM
Primary foetal cells were seeded across well plates for biochemical, histological and metabolic analysis or tissue culture flasks for microarray studies. After 24 hrs, cells were washed twice with PBS then given either CDM with activin A/FGF-2 (CDM + A/F) or basal medium to establish culture conditions. Media changes were performed on respective cultures every 48 hrs, and after 5 days CDM + A/F cultures were refreshed with either CDM alone (still labelled as CDM + A/F), CDM with 100 μM ascorbate/10 nM dexamethasone, CDM with 150 ng/ml BMP-2 or basal medium with basal medium changes on control wells. Concentrations of modulatory factors were used as established [12, 20]. Cultures were maintained for a further 5 days with media changes every 48 hrs prior to harvest after a total of 10 days in CDM or control conditions (culture regimen is presented diagrammatically in Fig. S1).
Biochemical analysis
Following fixation in 95% ethanol, cell lysate in 0.05% TRITON®-X100 was utilized for colorimetric turnover of para-Nitrophenylphosphate (pNPP), measured at 410 nm to quantify ALP activity as nmol pNPP/hr or assayed using the PicoGreen® double-stranded DNA quantification reagent (480 nm excitation, 520 nm emission) to calculate DNA content, given as ng/ml. Specific ALP activity results were expressed as nmol pNPP/ng DNA/hr. Absorbance and fluorescence were measured on an ELX-800 Universal Microplate Reader and FLX-800 Microplate Fluorescence Reader, respectively, using Bio-Tek KC4 Kineticalc for Windows software (version 3.01, revision 7) (Bio-Tek Instruments, Inc., Winooski, VT, USA, http://www.bio-tek.com). Values are expressed as mean ± standard deviation (S.D.). Studies were run in triplicate with multiple culture wells (n= 6). Statistical significance compared to basal control cultures was calculated using anova with Tukey’s Multiple Comparisons post-test. All statistical analyses were performed with GraphPad Instant Software (GraphPad Software Inc, San Diego, CA, USA, http://www.graphpad.com).
Metabolic activity
Following washes in PBS, samples were incubated in Earle’s balanced salt solution supplemented with 0.5% (v/v) human serum albumin, 1 mM glucose, 5 mM lactate, 0.47 mM pyruvate and a complete mixture of amino acids, for 2 hrs at 37°C prior to fixation. Reverse phase high performance liquid chromatography was performed as previously described [33]. Following correction for blank wells (media in the absence of cells), amino acid data were expressed as either appearance or depletion of amino acids from the culture medium, or the sum of the two given as ‘turnover’, expressed as pmol/ng DNA/h, mean ± standard error of the mean (S.E.M.). Amino acid data were analysed to determine whether they were normally distributed using the Ryan-Joiner normality test. Differences between basal conditions and CDM with activin A and FGF-2 were analysed using either a Student’s t-test or a Mann-Whitney U-test. Differences between CDM with activin A/FGF-2, CDM with ascorbate/dexamethasone and CDM with BMP-2 were analysed using one-way anova followed by a Fisher’s test.
Cell viability
Cells were labelled with Cell Tracker Green™ CMFDA and ethidium homodimer-1 for viable or necrotic cells, respectively, as per manufacturer’s instructions. Following fixation in 95% ethanol, cells were counterstained with 4′,6-diamidino-2-phenylindole (DAPI, 1:100 in PBS) for 5 min.
Fluorescence immunocytochemistry
Cultures were fixed in 4% paraformaldehyde and stored in PBS at 4°C prior to immunostaining. Following blocking (1% BSA in PBS) and permeablization where necessary (0.01% TRITON®-X100) primary antibodies were incubated at 4°C overnight with respective dilutions in 1% BSA in PBS; type I collagen (LF67), 1:300; type II collagen, 1:1000; STRO-1, 1:2; SOX-2, 1:200 or SOX-9, 1:150. Following PBS washes appropriate secondary antibodies were applied for 1 hr at room temperature and counterstained with DAPI. Negative controls lacked the primary antibody, wherein no staining was observed (Fig. S2).
RNA extraction and cDNA synthesis
Total RNA was extracted from cell cultures using the TRIzol reagent as per manufacturer’s instructions and subjected to DNAse treatment then either sent for microarray analysis or reverse transcribed using the super-script first-strand synthesis system for PCR.
Microarray analysis
Microarray processing was conducted by Precision Biomarker Resources, Inc., (Evanston, IL, USA, http://www.precisionbiomarker.com). Following confirmation of sample quality on a nanodrop ND-1000 (for optical densities at 230, 260 and 280) and a Bioanalyzer Nanochip (Agilent’s protocol), 1.0 μg of labelled cRNA from each sample was hybridized to Affymetrix U133 plus 2.0 chips in triplicate (giving three technical replicates per sample, with three biological replicates) and processed using the Affymetrix GeneChip Array Station (GCAS) protocol according to manufacturer’s instructions. To determine significantly altered gene expression for basal medium versus CDM with activin A/FGF-2, a pairwise Student’s t-test with a Bonferroni correction (P < 0.05/length or array, or 54,675 probe sets) was performed from log2 normalized expression data with MATLAB® software (version 7.3, The Mathworks Inc., Natick, MA, USA, http://www.mathworks.com) (Fig. S3A). A three-way anova was performed to compare basal, CDM with activin A/FGF-2 and CDM plus BMP-2 conditions by Dr. Eric Bremer of Precision Biomarker Resources, Inc. using Partek® Genomics Suite software (version 6.08, Partek Inc., St. Louis, MO, USA, http://www.partek.com). Significantly altered gene expression (P < 0.05) above a twofold threshold was determined by the G mean ratio of log2 normalized expression data (Fig. S3B). Appropriate genes of interest for qPCR validation were determined following literature review. Microarray data were deposited in Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) and Array Express (http://www.ebi.ac.uk/arrayexpress/) in accordance with MIAME guidelines [34].
Quantitative PCR
Real-time qRT-PCR was performed with the Applied Biosystems 7500 Real Time PCR System for primers as illustrated in Table 2. Values were calculated using the comparative threshold cycle (Ct) method and normalized to β-ACTIN expression. Results of combined experiments have been presented (three donor samples) and expressed as mean ± S.D. Statistical analyses were performed with Student’s t-test for CDM and activin A/FGF-2 cultures in comparison to basal conditions or one-way anova with Tukey’s multiple comparisons post-test for basal, CDM with activin A/FGF-2 and CDM with BMP-2 comparisons.
| Gene abbr. | Full name | Accession number | Primer sequences | Amplicon |
|---|---|---|---|---|
| β-Actin | β-actin | NM_001101 | F: 5′ ggc atc ctc acc ctg aag ta 3′ | 81 bp |
| R: 5′ agg tgt ggt gcc aga ttt tc 3′ | ||||
| ALP | Alkaline phosphatase | NM_000478 | F: 5′ gga act cct gac cct tga cc3′ | 85 bp |
| R: 5′ tcc tgt tca gct cgt act gc 3′ | ||||
| COL1A1 | Collagen type I, α1 | NM_000088 | F: 5′ aac agc cgc ttc acc tac ag 3′ | 99 bp |
| R: 5′ ggg agg tct tgg tgg ttt tg 3′ | ||||
| OCN | Bone γ-carboxyglutamate protein (osteocalcin) | NM_199173 | F: 5′ ggc agc gag gta gtg aag ag 3′ | 101 bp |
| R: 5′ ctc aca cac ctc cct cct g 3′ | ||||
| RUNX2 | Runt-related transcription factor 2 | NM_001015051 | F: 5′ tct tca caa atc ctc ccc 3′ | 230 bp |
| R: 5′ tgg att aaa agg act tgg tg 3′ | ||||
| CLU | Clusterin | NM_001831 | F: 5′ cca gac ggt ctc aga caa tg 3′ | 95 bp |
| R: 5′ gtt tca ccc cgt tga cag 3′ | ||||
| CRYAB | Crystallin, αb | NM_001885 | F: 5′ tgg gag atg tga ttg agg tg 3′ | 81 bp |
| R: 5′ tcc tgt gga act ccc tgg 3′ | ||||
| CSRP1 | Cysteine and glycine-rich protein 1 | NM_004078 | F: 5′ gct ggg tat caa gca cga g 3′ | 91 bp |
| R: 5′ ctc gga gcc acc aat ctt ct 3′ | ||||
| EPAS1 | Endothelial PAS domain protein 1 | NM_001430 | F: 5′ aac ctc aag tca gcc acc tg 3′ | 75 bp |
| R: 5′ gtg agg agg gca gtt gtt gt 3′ | ||||
| GREM1 | Gremlin 1 | NM_013372 | F: 5′ cac act caa ctg ccc tga a 3′ | 73 bp |
| R: 5′ gca acg aca ctg ctt cac 3′ | ||||
| MT1X | Metallothionein 1X | NM_005952 | F: 5′ caa ctg ctc ctg ctc gcc 3′ | 104 bp |
| R: 5′ ggc agc agg agc agc agc 3′ | ||||
| OSR2 | Odd-skipped related 2 | NM_053001 | F: 5′ gtg aca tct gcc aca agg 3′ | 104 bp |
| R: 5′ tcc ttt ccc aca ctc ctg 3′ | ||||
| POSTN | Periostin, osteoblast specific factor | NM_006475 | F: 5′ gca ccg agt aat gag gct tg 3′ | 61 bp |
| R: 5′ tgc tct cca aac ctc tac gg 3′ | ||||
| RABGAP1 | RAB GTPase activating protein 1 | NM_012197 | F: 5′ ggg ctc caa aaa cca gag 3′ | 83 bp |
| R: 5′ gcc act ggt gtg aaa gga 3′ | ||||
| SRGN | Serglycin | NM_002727 | F: 5′ ccg tct gag gac tga cct tt 3′ | 93 bp |
| R: 5′ ccg aag cct gat cca gag ta 3′ | ||||
| C1orf19 | Chromosome 1 open reading frame 19 | NM_052965 | F: 5′ gtt tac ctg gac ctc atg g 3′ | 86 bp |
| R: 5′ cca aca agg cag atg agc 3′ | ||||
| CHSY-2 | Chondroitin synthase-2 | NM_175856 | F: 5′ cgc cga cga cga tgt cta c 3′ | 85 bp |
| R: 5′ tcc cag gta gag agg ctt act g 3′ | ||||
| DUSP6 | Dual specificity phosphatase 6 | NM_022652 | F: 5′ ggg caa gaa ctg tgg tgt ct 3′ | 59 bp |
| R: 5′ cag tga ctg agc ggc taa tg 3′ | ||||
| GFPT2 | Glutamine-fructose-6-phosphate transaminase 2 | NM_005110 | F: 5′ cgg ctg gag tac aga ggc ta 3′ | 99 bp |
| R: 5′ ccc cct ttt ctt gac cag 3′ | ||||
| HMGCS1 | 3-hydroxy-3-methylglutaryl-Coenzyme A synthase 1 | NM_001098272 | F: 5′ cgt ccc act cca aat gat g 3′ | 94 bp |
| R: 5′ ctt tct tgg cag ggc ttg 3′ | ||||
| PPL | Periplakin | NM_002705 | F: 5′ gga ggc act ctg tga ctt tg 3′ | 84 bp |
| R: 5′ ctc tcc ccg ttg ttc ttc tg 3′ | ||||
| SC4MOL | Sterol-C4-methyl oxidase-like | NM_001017369 | F: 5′ cat ggg tga cca ttc gtt tat 3′ | 146 bp |
| R: 5′ tg aag cat agt ttc caa tga agt t 3′ | ||||
| SOX15 | SRY (Sex determining region Y)-box 15 | NM_006942 | F: 5′ aac tgc tgc cca cct ata cc 3′ | 65 bp |
| R: 5′ aag ggg agg gtt gta tgg ag 3′ | ||||
| SQLE | Squalene epoxidase | NM_003129 | F: 5′ gat ggg agt tca gta caa gg 3′ | 68 bp |
| R: 5′ caa cag tca gtg gag cat gg 3′ | ||||
| WIF1 | Wnt inhibitory factor 1 | NM_007191 | F: 5′ ctg ccc acc tgg att cta tg 3′ | 59 bp |
| R: 5′ aag cag gtg gtt gag cag 3′ |
Results
Human foetal femur cells grow in a serum-free chemically defined medium
To investigate the efficacy of the CDM, human foetal femur-derived cells were cultured in CDM with activin A/FGF-2 (Fig. 1). After 7 days, growth was observed to be equivalent to basal controls maintained in serum (Fig. 1A); however, the addition of activin A/FGF-2 to α-MEM alone resulted in a significantly reduced cell numbers as assessed by DNA content, (Fig. 1A) and negligible ALP activity (Fig. 1B). Cultures containing serum displayed a baseline level of ALP activity which was 10-fold greater than cultures with CDM and activin A/FGF-2 (Fig. 1B). Following genome-wide expression array analysis, 46 genes were identified as significantly different in three different foetal samples (Table S1). Down-regulated genes included matrix metalloproteinases (MMPs) 1, 3 and 13, the chemokine (C-X-C motif) ligands 1 and 6, interleukins 6 and 8 metallothionein variants. Validated expression by qPCR demonstrated up-regulation of clusterin, odd-skipped related 2, periostin and RAB GTPase 1 (Fig. 1C) and a loss of expression for crystallin αB, cysteine and glycine rich protein 1, endothelial PAS protein 1, gremlin 1, metallothionein 1X and serglycin (Fig. 1D). Metabolic analysis demonstrated significantly reduced amino acid depletion, appearance and turnover in cultures of CDM with activin A and FGF-2 compared to basal controls (Fig. 1E).
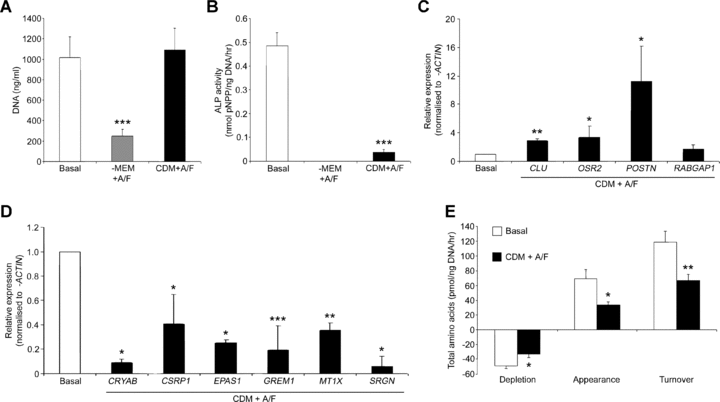
Culture of human foetal femur-derived cells in a serum-free chemically defined medium (CDM). (A) Cell number in populations maintained for 7 days in either α-MEM with 10% foetal calf serum (Basal), α-MEM with 10 ng/ml activin A and 12 ng/ml FGF-2 (α-MEM + A/F) or CDM with activin A/FGF-2 (CDM + A/F) as given by PicoGreen® double-stranded DNA assay. (B) Specific alkaline phosphatase (ALP) enzymatic activity for three treatment groups, (C) Up-regulation of CLU, OSR2, POSTN and RABGAP1 and (D) Down-regulation of genes CRYAB, CSRP1, EPAS1, GREM1, MT1X and SRGN in CDM plus activin A and FGF-2 cultures compared with basal cultures as validated by quantitative PCR from genome-wide expression array analysis. (E) Total amino acid depletion, appearance and turnover by foetal cells cultured under basal conditions or in CDM in the presence of activin A and FGF-2. Results expressed as mean ± S.D. (A–D) or mean ± S.E.M. (E) with n= 6 (A, B, E) or n= 3 (C, D); *P < 0.05, **P < 0.01, ***P < 0.001 significantly different from basal conditions. Transcript levels were determined using the comparative threshold cycle (Ct) method and normalized to β-ACTIN expression. Transcript levels have been represented on the y-axis relative to basal conditions for each cell population, set as 1. Abbreviations: α-MEM, α-modification minimum essential medium; CLU, clusterin; CRYAB, crystallin, αb; CSRP1, cysteine and glycine-rich protein 1; EPAS1, endothelial PAS domain protein 1; FGF-2, fibroblast growth factor 2; GREM1, gremlin 1; MT1X, metallothionein 1X; OSR2, odd-skipped related 2; PCR, polymerase chain reaction; pNPP, p-Nitrophenylphosphate; POSTN, periostin; RABGAP1, RAB GTPase activating protein 1; SRGN, serglycin.
Use of CDM to model osteogenic modulatory factors in human foetal cell cultures
The addition of 100 μM ascorbate and 10 nM dexamethasone to basal medium enhanced ALP activity in comparison to basal controls cultures (Fig. S4). No increase of ALP was observed with 150 ng/ml BMP-2, and, furthermore, supplementation of BMP-2 to ascorbate/dexamethasone-treated cultures attenuated their ALP activity (Fig. S4). We therefore modelled the effect of osteogenic modulatory factors on cultures in CDM (Fig. 2). In comparison to basal cultures, cells cultured in CDM supplemented with either activin A/FGF-2, ascorbate/dexamethasone or BMP-2 displayed reduced cell number, according to DNA content, which was restored to control levels when transferred back to basal culture conditions (Fig. 2A). The addition of ascorbate/dexamethasone resulted in only partial recovery of ALP activity in comparison to basal cultures (Fig. 2B). Moreover, the lowest specific ALP activity was observed in cultures grown in CDM plus BMP-2 (Fig. 2B). Morphologically, negligible difference was observed between basal conditions and CDM cultures containing either activin A/FGF-2, ascorbate/dexamethasone or medium with serum (Fig. 2C–F). In contrast, a marked change was observed in cultures supplemented with BMP-2 with appearance of a cobblestone-like cellular morphology, surrounded by cells with typical fibroblast morphology (Fig. 2G compared with Fig. 2C–F). Nuclear counterstaining demonstrated a shift in nuclear location to the cell edge and a kidney-shaped morphology (Fig. 2H and inset). To determine culture viability, CDM plus BMP-2 conditions were maintained for 21 days with negligible evidence of cell death (Fig. 2I and J). Metabolic studies showed significantly increased amino acid depletion, appearance and turnover in CDM with ascorbate/dexamethasone and CDM with BMP-2 cultures in comparison to CDM and activin A/FGF-2 (Fig. 2K).
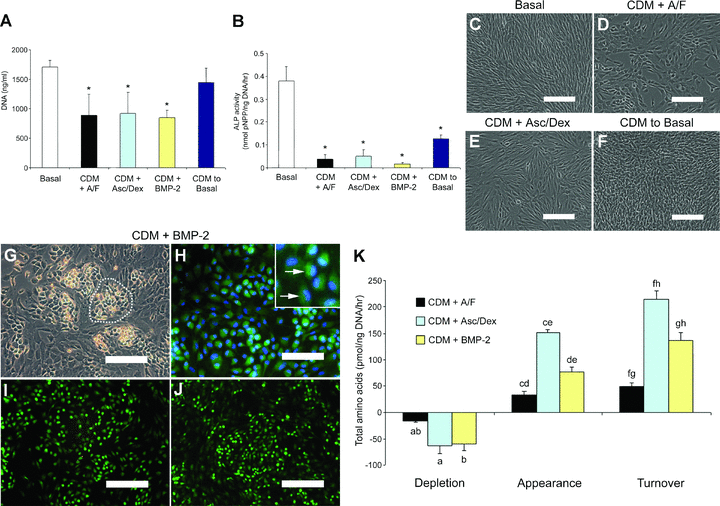
Addition of osteogenic factors to chemically defined medium (CDM)-treated human foetal femur-derived cell cultures. (A) PicoGreen® double-stranded DNA assay gave cell number for day 10 cultures of CDM with 10 ng/ml activin A and 12 ng/ml FGF-2 (CDM + A/F), CDM with 100 μM ascorbate/10 nM dexamethasone (CDM + Asc/Dex), CDM with 150 ng/ml BMP-2 (CDM + BMP-2) or cultures maintained in CDM with activin A and FGF-2 for 5 days and transferred to basal medium for a final 5 days (CDM to Basal). (B) Specific alkaline phosphatase (ALP) activity of culture groups in comparison to basal controls. (C) Typical fibroblastic morphology of basal cultures, also seen in; (D) CDM with activin A and FGF-2, (E) CDM with ascorbate/dexamethasone and; (F) cells pre-treated with CDM plus activin A and FGF-2 before returning to basal conditions. (G) Human foetal cells displayed colonies of cobblestone-like cells, in day 10 CDM cultures with BMP-2 (example denoted by dotted line). (H) CDM plus BMP-2 day 10 cultures labelled with Cell Tracker Green™ CMFDA and DAPI nuclear counterstain. INSET: Cobblestone-like cells and kidney-shaped nuclei (arrows). (I) Cell Tracker Green™ CMFDA and ethidium homodimer-1 (red) labelling for live and necrotic cells, respectively, in CDM plus BMP-2 cultures at day 14 (J) and at day 21. (K) The effect of different culture components on total amino acid depletion, appearance and turnover by foetal femur cells. Bars = 200 μm (C–G, I, J), 100 μm (H). Results expressed as mean ± S.D. (A, B) or mean ± S.E.M. (K), n= 6. *P < 0.001 significantly different from basal conditions (A, B); bars with the same superscript are significantly different; a, b, P < 0.05; c, d, e, f, g, h, P < 0.001 (K). Abbreviations: BMP-2, bone morphogenetic protein-2; CMFDA, 5-chloromethyl-fluorescein diacetate; DAPI, 4′,6-diamidino-2-phenylindole; FGF-2, fibroblast growth factor 2; pNPP, para-Nitrophenolphosphate.
Phenotypic characterization demonstrated a loss of type I collagen expression in both CDM with BMP-2 and CDM plus activin A/FGF-2 conditions in comparison to basal cultures (Fig. 3AversusFig. 3B and C), with minimal expression of type II collagen (Fig. 3DversusFig. 3E and F). STRO-1 was seen in selected cells in three conditions (Fig. 3G–I) although not in the cells with the altered nuclear positioning (Fig. 3I). SOX-9 expression was observed in basal (Fig. 3J) and CDM with activin A/FGF-2 cultures (Fig. 3K) but limited to the cobblestone-like cells in CDM plus BMP-2 (Fig. 3L). In addition, expression of SOX-2 was observed in the CDM plus BMP-2 cultures only and was restricted to the cobblestone-like cells (Fig. 3M and N versusFig. 3O). Expression of OCT4 and TRA-1–60 were not seen in any of the treatments (Fig. S5).
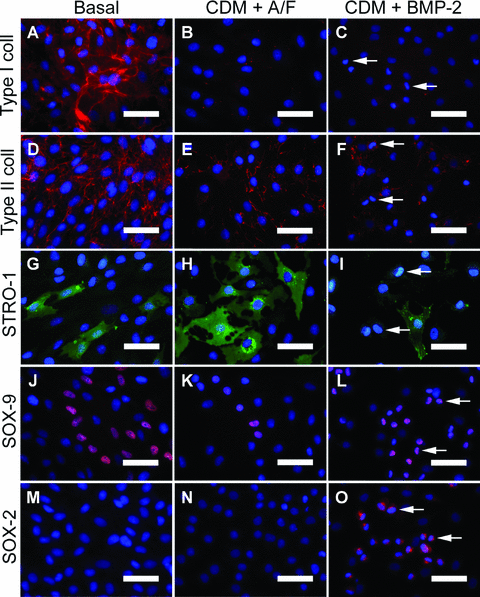
Expression of mesenchymal and progenitor markers in human foetal femur-derived cells maintained with either basal medium containing serum (Basal), serum-free chemically defined medium with 10 ng/ml activin A and 12 ng/ml FGF-2 (CDM + A/F) or CDM containing 150 ng/ml BMP-2 (CDM + BMP-2). (A–C) Fluorescence immunocytochemistry staining for type I collagen in respective cultures, as for; (D–F) type II collagen, (G–I) STRO-1, (J–L) SOX-9 and (M–O) SOX-2. Cobblestone-like cells in CDM plus BMP-2 cultures denoted by arrows (C, F, I, L, O). Bars = 50 μm. Abbreviations: BMP-2, bone morphogenetic protein-2; FGF-2, fibroblast growth factor 2.
Microarray analysis of CDM plus BMP-2 cultures
To better define the mechanisms behind the altered phenotype in the CDM plus BMP-2 cultures, genome-wide expression array analysis was used to identify differential gene expression compared to basal and CDM with activin A/FGF-2 conditions (Fig. 4). Principal component analysis of genome-wide expression arrays showed a distinct shift in the dimensionality of gene clustering, indicating a markedly altered gene expression profile in cells cultured in CDM with BMP-2 compared to basal or CDM plus activin A/FGF-2 conditions (Fig. 4A). Results of a three-way anova comparing the culture conditions demonstrated significantly altered expression of 58 genes and hierarchical cluster analysis showed basal conditions as more distinct to either CDM treatment group as shown by the length of the vertical dendrogram arms, with little variation between replicates or samples (Fig. 4B). Ten genes were identified as representative from the six cluster groups formed at the fourth level of horizontal dendrogram arms and validated with qPCR (Fig. 5). Expression levels for chromosome 1 open reading frame 19 increased in CDM with activin A/FGF-2 cultures over basal and was higher still in CDM plus BMP-2 conditions (Fig. 5A), a pattern repeated for 3-hydroxy-3-methylglutaryl-coenzyme A synthase 1 (Fig. 5B), sterol-C4-methyl oxidase-like (Fig. 5C), and Wnt inhibitory factor 1 (Fig. 5D). The highest expression for dual specificity phosphatase 6, periplakin and SOX15 were observed in CDM with activin A/FGF-2 conditions, compared to basal and CDM plus BMP-2 cultures (Fig. 5E–G, respectively). In CDM plus BMP-2 conditions, expression of chondroitin synthase-2 was comparable to basal medium (Fig. 5H) but had the highest expression of glutamine-fructose-6-phosphate transaminase 2 (Fig. 5I) and squalene epoxidase (Fig. 5J) in comparison to the other two treatment groups.
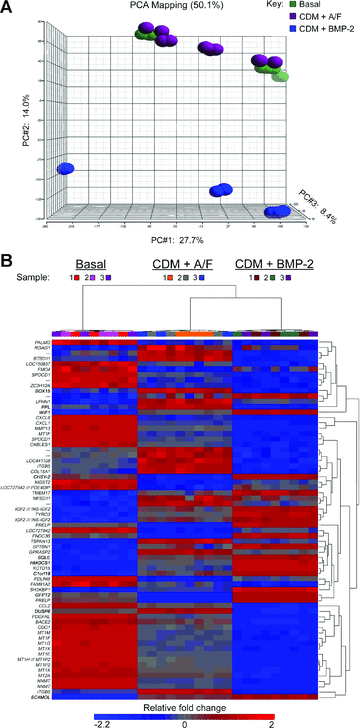
Genome-wide expression array analysis for basal medium containing serum (Basal), serum-free chemically defined medium (CDM) with 10 ng/ml activin A and 12 ng/ml FGF-2 (CDM + A/F) or CDM containing 150 ng/ml BMP-2 (CDM + BMP-2). (A) Principal component analysis (PCA) 3D scatter plot for three foetal tissue samples hybridized to genome wide expression array chips showing clusters of replicate technical samples and shift of cluster points in cells cultured in CDM in the presence of BMP-2 in comparison to cells maintained in either basal medium or CDM supplemented with activin A and FGF-2. (B) Heat map of significantly differently expressed genes in human foetal femur-derived cells cultured in basal, CDM with activin A/FGF-2 or CDM plus BMP-2 conditions. Results of the three-way anova comparing basal medium (containing serum), serum-free CDM containing 10 ng/ml activin A and 12 ng/ml FGF-2 or CDM with 150 ng/ml BMP-2 determined 58 genes, with a fold change ranging between –2.2 and 2. Hierarchical cluster analysis shows basal conditions as more distinct to either CDM treatment group as shown by the length of the vertical dendrogram arms, with little variation between replicates or samples. Ten genes of interest, shown in bold, were identified from the six cluster groups formed at the fourth level of horizontal dendrogram arms: Chromosome 1 open reading frame 19 (C1orf19); chondroitin synthase-2 (CHSY-2); dual specificity phosphatase 6 (DUSP6); glutamine-fructose-6-phosphate transaminase 2 (GFPT2); 3-hydroxy-3-methylglutaryl-coenzyme A synthase 1 (HMGCS1); periplakin (PPL); sterol-C4-methyl oxidase-like (SC4MOL); SRY (sex determining region Y)-box 15 (SOX15); squalene epoxidase (SQLE) and Wnt inhibitory factor 1 (WIF1). n= 3 per treatment group as shown by key. Abbreviations: BMP-2, bone morphogenetic protein-2; FGF-2, fibroblast growth factor 2.
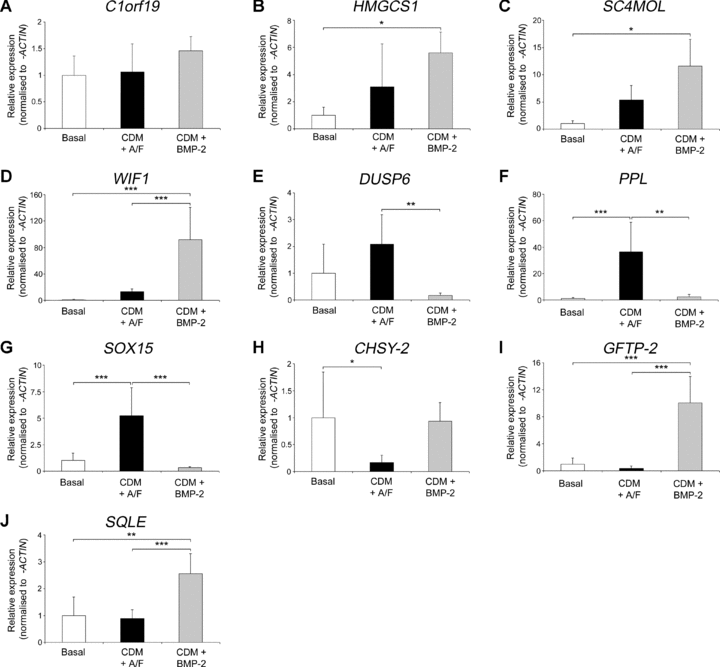
Quantitative PCR validation of significantly altered gene expression between basal and serum-free treatment groups. Foetal cell cultures were maintained in either basal medium with serum (Basal), CDM with 10 ng/ml activin A and 12 ng/ml FGF-2 (CDM + A/F) or CDM plus 150 ng/ml BMP-2 conditions(CDM + BMP-2) and gene expression measured for; (A) Chromosome 1 open reading frame 19 (C1orf19), (B) 3-hydroxy-3-methylglutaryl-coenzyme A synthase 1 (HMGCS1); (C) Sterol-C4-methyl oxidase-like (SC4MOL), (D) Wnt inhibitory factor 1 (WIF1), (E) Dual specificity phosphatase 6 (DUSP6), (F) Periplakin (PPL), (G) SRY (sex determining region Y)-box 15 (SOX15), (H) Chondroitin synthase-2 (CHSY-2), (I) Glutamine-fructose-6-phosphate transaminase 2 (GFPT2) and (J) Squalene epoxidase (SQLE). Transcript levels were determined using the comparative threshold cycle (Ct) method and normalized to β-ACTIN expression with basal conditions set as 1. Results expressed as mean ± S.D. with n= 3; *P < 0.05, **P < 0.01, ***P < 0.001. Abbreviations: BMP-2, bone morphogenetic protein-2; FGF-2, fibroblast growth factor 2, PCR, polymerase chain reaction.
Discussion
We have utilized a serum-free CDM for human foetal femur cell expansion and demonstrated the derivation of an undifferentiated population compared to cultures maintained in serum. Furthermore, the addition of BMP-2 provides a novel cellular model for differentiation studies.
These studies highlight several key findings; firstly, culture in CDM with activin A and fibroblast growth factor-2 (FGF-2) [6, 9] resulted in an undifferentiated phenotype as indicated by reduced amino acid turnover [35], negligible expression of ALP activity and type I and type II collagens. Expression of STRO-1 and SOX-9, as markers of mesenchymal progenitors, was maintained in selected cells [36, 37]. Microarray studies confirmed this phenotype, with up-regulation of clusterin, involved in anti-apoptosis [38, 39], odd-skipped related 2 and periostin, as key regulators of pre-osteoblast proliferation [40, 41] and RAB GTPase 1, indicated in cell cycle maintenance [42]. In addition, we observed down-regulation of crystallin αB, suggested to have a role in osteogenic differentiation [43], gremlin 1, a BMP antagonist [44] and cysteine and glycine-rich protein 1, involved in the regulatory processes of development and cellular differentiation [45] and implicated in the non-canonical Wnt signalling pathway [46]. Down-regulated metallothioneins correlate to studies which showed a positive regulatory role of metallothionein isoforms in osteogenesis [47] and reduced serglycin expression with CDM and activin A/FGF-2 suggests a role in the proliferative capacity of these cells [48]. Further genes down-regulated include interleukin 6 (IL-6), necessary for osteoblast signalling in osteoclastogenesis [49] and chemokine C-X-C motif ligands 1 and 6, seen in early neural progenitors during development of the human foetal brain [50] and in mesenchymal cells [51], respectively. Down-regulation of MMPs 1, 3 and 13, confirmed the de-differentiation effects of serum-free CDM with activin A/FGF-2, with the latter isoform seen in hypertrophic chondrocytes of human foetal bone tissue [52], similar in locus and morphology to the foetal femur tissue utilized in the current studies [31].
Additionally, although the application of ascorbate/dexamethasone to adult and foetal mesenchymal populations has been previously reported [12, 53] and BMP-2 has been shown to positively affect hBMSC proliferation [20, 22, 54] the current study demonstrated the addition of BMP-2 resulted in a significant reduction of ALP activity and foetal cell number. Moreover, culture in CDM with BMP-2 resulted in a marked change to colonies of cobblestone-like cells supported by typical fibroblastic cells, and although cobblestone-like cells displayed an altered nuclear location and shape; the cells maintained viability in extended culture. Reduced expression of the cell adhesion protein periplakin [55], in comparison to cultures with activin A/FGF-2, may be linked to the cobblestone-like morphology. Although STRO-1+ cells were observed with BMP-2, these were not the cobblestone-like cells. Conversely, SOX-9 expression was restricted to the cobblestone-like cells only, as was SOX-2, a recognized stem cell marker [56], although the extranuclear expression may be linked to the altered nuclear shape. In contrast to the study of foetal mesenchymal cells by Guillot and coworkers [57], expression of other hESC markers OCT4 and TRA-1–60 [7, 58, 59] were not seen in the human foetal femur-derived cells. These cultures were further characterized by an increased metabolic activity in comparison to CDM and activin A/FGF-2 conditions, however, suggestive of a differentiating population [33, 60], as supported by a marked increase in the expression of metabolic gene glutamine-fructose-6-phosphate transaminase 2 [61] and two markers of sterol biosynthesis – sterol-C4-methyl oxidase-like [62] and squalene epoxidase [63]. Up-regulation of the tRNA splicing endonuclease 15 homolog chromosome 1 open reading frame 19 suggests these populations may also utilise extra mechanisms to maintain cell growth [64]. Down-regulation of differentiation regulatory genes was confirmed with SOX15, involved in skeletal muscle development [65], and DUSP6, a regulator of the MAP kinase pathway involved osteogenic signal transduction [66]. In addition, increased expression of Wnt inhibitory factor 1, a major antagonist of BMP-signalling [67], was seen in CDM cultures with BMP-2, as was the synthesizing enzyme 3-hydroxy-3-methylglutaryl-coenzyme A synthase 1, part of the statins family previously indicated in osteogenesis [68].
FGF-2, a developmental mitogen, has been shown to preferentially target primary and precursor cell populations for the expansion of chondro- and osteoprogenitors [69–74]. Interestingly, Martin and coworkers suggested FGF-2 may result in a de-differentiation of committed mesenchymal progenitors prior to the addition of BMP-2 to promote lineage differentiation [75], in contrast however to the current studies. FGF-2 signalling may be mediated by chondroitin sulphate glycosaminoglycans (CS-GAGs) [76]via a recently proposed mechanism whereupon release from a CS-GAG-protected extracellular matrix environment exposes a newly divided daughter cell to exogenous growth factors [77]. We observed increased levels of chondroitin synthase-2 (also known as chondroitin sulphate synthase 3) suggesting that BMP-2 maintains this ‘protective’ environment in these studies. Furthermore, FGF-mediated induction of SoX2 expression in murine osteoblasts indicated FGF regulation of the Wnt/ β-catenin pathways [79] and co-operation of TGF-β/BMP-2 and Wnt signalling pathways has been demonstrated to enhance chondrogenesis [80–82]. The human foetal femur-derived cells are derived initially from predominantly cartilaginous explants [31] and widespread SOX-9 expression in the native femur tissue may be indicative of a mesenchymal condensation, an upstream event of chondrogenesis [37, 83]. BMPs promote transient SOX gene expression influential in the terminal differentiation of chondrocytes [21] and in serum-free CDM conditions may therefore promote a chondrogenic response in the human foetal cells through the selection of a sub-population of cells or early progenitors. This is supported by the study of Schmitt and coworkers who observed differentiation along the chondrogenic, but not osteogenic or adipogenic lineages, in hBMSCs in a serum-free environment with BMP-2 [78]. That the cobblestone-like cells alone expressed SOX-9 and SOX-2, but not STRO-1 or type I and II collagens suggested a differential response of sub-populations to BMP-2. These findings indicate a novel in vitro model for the recapitulation of the condensation phenotype, in the presence of CDM and ongoing work in our laboratories is focused on functionality of cobblestone-like cell preparations.
Finally, a key consideration for further serum-free CDM studies is the potential for osteogenic rescue. However, the use of serum-free CDM remains an important tool for the ex vivo expansion of human mesenchymal cells [84]. Moreover, CDM conditions may prove more appropriate for mesenchymal expansion as the latent osteogenic activity seen in the human foetal femur-derived cells in standard basal culture conditions demonstrates competency factors within serum, with implications for in vitro expansion and osteogenic studies, namely (i) additional factors may be necessary for osteogenic differentiation, as; (ii) osteogenesis may not be the result of added modulatory factors alone and (iii) peripheral differentiation pathways may therefore be overlooked.
In conclusion, the current studies confirm the potential of CDM-mediated ex vivo expansion of human foetal femur-derived cells and the derivation of a unique undifferentiated phenotype in the presence of activin A and FGF-2. The primitive foetal populations generated with BMP-2 offer a new model to investigate the underlying molecular mechanisms in skeletal stem and progenitor cell biology with implications for tissue regeneration strategies.
Acknowledgements
The authors would like to thank Prof. David I. Wilson for his support in securing foetal samples. The polyclonal antibody to type I collagen was a generous gift from Dr. Larry Fisher (NIH). N.A.H. is a U.K. Department of Health Clinician Scientist We also thank Dr. Eric Bremer of Precision Biomarker Resources, Inc. for assistance with microarray anova studies and Catherine Forristal for provision of hES cells for control staining. This work was funded by the Biotechnology and Biological Sciences Research Council, the U.K. Department of Trade and Industry through the Technology Strategy Board, the Wellcome Trust, the Engineering and Physical Sciences Research Council and the Research Management Committee of the School of Medicine, University of Southampton.