A TEST FOR ENVIRONMENTAL EFFECTS ON BEHAVIORAL ISOLATION IN TWO SPECIES OF KILLIFISH
Abstract
Behavioral isolation is a common and potent mechanism of reproductive isolation. Determining the extent to which behavioral isolation varies with environmental conditions is critical to understanding speciation and the maintenance of species boundaries. Here, we tested the effect of salinity on behavioral isolation (female species recognition, male–male competition, and male species recognition) between two closely related killifish (Lucania goodei and L. parva) that differ in salinity tolerance. We performed no-choice assays and behavioral trials where males could compete and court females in fresh water (0 ppt) and brackish water (15 ppt). We found high levels of behavioral isolation that did not vary as a function of salinity. In behavioral trials, male species recognition of females was strong and asymmetric between the two species. Lucania goodei males preferred conspecifics and rarely courted or mated with L. parva females. Lucania parva males preferred conspecifics but readily courted and mated with L. goodei females. This asymmetry matches previously documented asymmetries in hybrid offspring fitness. Crosses between L. parva males and L. goodei females produce fully viable/fertile hybrids, but crosses between L. goodei males and L. parva females produce males with reduced fertility. Hence, behavioral isolation may have evolved in part due to reinforcement.
Behavioral isolation, where individuals prefer to mate with members of their own population/species, is one of the most common forms of reproductive isolation in animals (Mayr 1963; Coyne and Orr 2004; Ritchie 2007). The critical task is determining how preferences diverge over time to produce behavioral isolation (Rundle and Boughman 2010). Behavioral isolation is generally thought to evolve via two main pathways. First, behavioral isolation may evolve as a consequence of divergent natural selection due to different environmental conditions (i.e., an ecological mechanism, Schluter 2001; Boughman et al. 2005; Rundle and Boughman 2010). Examples of this include (1) good genes sexual selection when the character trait is ecologically relevant, (2) sensory drive where environmental conditions affect signaling dynamics, and (3) reinforcement due to extrinsic isolation (Endler 1992; Boughman 2001; Boughman 2002; Servedio and Noor 2003; Rundle and Boughman 2010; Maan and Seehausen 2011). Second, behavioral isolation may evolve independently of divergent natural selection to different environmental conditions (a nonecological mechanism, Rundle and Boughman 2010; Schluter 2001). For instance, behavioral isolation may evolve via divergent sexual selection where the direction of selection is independent of environmental conditions. Examples of this include sexual conflict or runaway sexual selection where preference initially diverges due to chance events (Lande 1981; Kirkpatrick and Ryan 1991; Noor 1999; Arnqvist et al. 2000; Hall et al. 2000; Martin and Hosken 2003; Servedio and Noor 2003). Behavioral isolation may also evolve via reinforcement due to intrinsic isolation when low hybrid fitness creates selection for individuals to mate with conspecifics regardless of environmental conditions (Howard 1993; Noor 1999; Servedio and Noor 2003; Coyne and Orr 2004).
Although ecological and nonecological mechanisms can both lead to behavioral isolation between species, they have different implications for ecological population divergence (i.e., ecological speciation) in the face of gene flow. If a nonecological mechanism drives the evolution of behavioral isolation, then ecological adaptation and nonrandom mating are conceivably independent traits that require linkage disequilibrium in order for speciation to occur (Felsenstein 1981; Gavrilets 2004; Servedio 2009). Conversely, if an ecological mechanism drives the evolution of behavioral isolation, then the genes that confer behavioral isolation are also under ecological selection themselves. In this case, two separate sets of genes are under the same selection pressure that generates strong linkage disequilibrium between, them making ecological speciation more likely (Boughman 2001; Gavrilets 2004; Kirkpatrick and Ravigne 2002). Hence, population divergence and speciation is more feasible when behavioral isolation is ecologically dependent.
One sign of behavioral isolation evolving due to an ecological mechanism is environment-dependent behavioral isolation where species exert their strongest preferences for conspecifics in their natal environments. For instance, ornaments/signals that reflect local adaptation (as in good genes for ecologically relevant character traits) may be sensitive to the local environment (Andersson 1994; Badyaev and Snell-Rood 2006; van Doorn et al. 2009). In this case, traits involved in species recognition are plastic with respect to the natal environment and thus are only correctly expressed when animals are raised in their native habitats. Similarly, under sensory drive, sexual ornaments/signals that have been selected for high detection in a specific environment (Endler 1992; Endler and Basolo 1998; Boughman 2002; Slabbekoorn and Smith 2002; Patten et al. 2004; Badyaev and Snell-Rood 2006; Seehausen et al. 2008; Cocroft et al. 2010; Tobias et al. 2010) may not be expressed properly or may not be effective at stimulating the receiver's sensory system in nonnative or disrupted habitats (Long and Houde 1989; Milinski and Bakker 1990; Seehausen et al. 1997; Fisher et al. 2006; Lewandowski and Boughman 2008; Tobler et al. 2008; Plath et al. 2010).
Alternatively, behavioral isolation may evolve independently of ecological conditions. Certain forms of sexual selection—where the costs and benefits of preferences and their associated traits are independent of the environment—can lead to behavioral isolation (Lande 1981; Higashi et al. 1999; Arnqvist et al. 2000; Martin and Hosken 2003; Ritchie 2007). Similarly, reinforcement driven by intrinsic, genetic incompatibilities can also lead to behavioral isolation (Howard 1993; Servedio and Noor 2003). Reinforcement makes several testable predictions. It predicts that behavioral isolation is greater in areas of sympatry between two species (Noor 1999; Servedio and Noor 2003; Coyne and Orr 2004). This prediction has been tested and verified in a number of systems (Servedio and Noor 2003; Coyne and Orr 2004). Reinforcement also predicts that behavioral isolation will be asymmetric in species pairs with asymmetrical intrinsic isolation such that behavioral isolation will be greatest in the direction where hybrids have the lowest fitness (Hoskin et al. 2005; Jaenike et al. 2006; Yukilevich in press). This theory has been verbally stated (e.g., Hoskin et al. 2005; Jaenike et al. 2006) and shown theoretically by Yukilevich (in press). A recent meta-analysis of Drosophila showed that in sympatry 15 of 16 species pairs had asymmetrical prezygotic isolation that matched the direction of asymmetrical postzyotic isolation (Yukilevich in press). In contrast, only 10 of 20 allopatric pairs had matching directions of pre- and postzygotic isolation (Yukilevich in press).
STUDY SYSTEM
In this study, we test for environment-dependent behavioral isolation between two closely related species of killifish (Lucania goodei and L. parva) that differ in the salinity tolerance but still have appreciable geographic overlap. Lucania goodei and L. parva are two recently diverged sister species (Duggins et al. 1983; Whitehead 2010). Despite having approximately 98% sequence similarity (R. C. Fuller, unpubl. data), these two species differ radically in their salinity tolerance. Lucania goodei is found primarily in freshwater sites (restricted mainly to Florida and southern Georgia) whereas L. parva is euryhaline and can be found in fresh, brackish, and marine habitats as far north as Massachusetts and as far west as central Mexico (Lee 1980). Lucania parva is also found in a wider range of temperatures (8.8–37.5°C) than L. goodei (13.5–35°C) (Arndt 1971). The wider range of salinity tolerance and temperature tolerance exhibited by L. parva may have allowed them to inhabit a wider geographical range. Differences in population salinity correspond with differential adaptation to salinity at multiple life stages (Dunson and Travis 1991; Fuller et al. 2007; Fuller 2008a). Ancestral reconstruction of salinity tolerance suggests that the common ancestor of these two species was marine (Whitehead 2010).
Both species are extremely iteroparus and will lay eggs continuously on aquatic vegetation during the breeding season (April–September depending on local temperatures). Eggs of both species are small and clear and will incubate for five to 14 days depending on temperature (Arndt 1971; E. L. Berdan, pers. obs.). After hatching, fry of both species will reach sexual maturity in two to four months depending on temperature and density (Foster 1967; Arndt 1971; E. L. Berdan, pers. obs.).
The two species exhibit multiple forms of reproductive isolation including behavioral, extrinsic, and intrinsic isolation (Fuller 2008a). Intrinsic isolation between the two species is asymmetric where crosses between L. parva males and L. goodei females produce viable male and female F1 hybrids, whereas crosses between L. goodei males and L. parva females produce viable F1 hybrid females, but partially fertile F1 hybrid males. None of the reproductive isolating barriers are complete, and there is evidence for current gene flow between the two species (R. C. Fuller, unpubl. data; Hubbs et al. 1943). Approximately 15% of L. goodei sites and 17% of L. parva sites in Florida are sympatric (Fuller and Noa 2008). This is a severe underestimate for L. parva as its range extends northward along the Atlantic coast to Massachusetts and westward along the Gulf Coast to Mexico; L. goodei is absent from these areas (Lee 1980). Generally, allopatric L. goodei populations occur in the interior regions of Florida; allopatric L. parva populations occur along the coast; sympatric populations occur over large stretches where freshwater rivers meet the coast (Fuller and Noa 2008). Rivers with high levels of dissolved ions also have many sympatric populations. When sympatric, the two species likely compete because food preferences of both species are similar (Arndt 1971).
We tested whether behavioral isolation is environment-dependent in relation to salinity. Salinity can alter the signaling dynamics in chemical communication systems of many organisms by influencing chemical signal detection (Sola and Tongiorgi 1996; Herbert-Read et al. 2010) and chemoreceptor properties (Gleeson et al. 1996). Salinity also affects multiple gene expression and hormone pathways that are likely to influence behavior (Sakamoto et al. 2001; Kitano et al. 2010). We measured several components of behavioral isolation (female species recognition, male-male competition, and male species recognition) between L. goodei and L. parva and determined whether they varied with salinity. If behavioral isolation has been driven by adaptation to salinity (i.e., via an ecological mechanism), we predict that both species will exert their strongest preferences for conspecifics at their preferred salinity (freshwater for L. goodei, saltwater for L. parva). Additionally, we wanted to determine if behavioral isolation is particularly strong in our system relative to estimates from other taxa. We calculated the IPSI statistic to estimate behavioral isolation and compared it to other IPSI estimates from the literature. We also calculated the overall and relative contributions of behavioral isolation and other reproductive isolating barriers using the methods of Ramsey et al. (2003).
Methods
EXPERIMENTAL ANIMALS
Adult L. parva were collected from Indian River Lagoon near Titusville (Brevard County, Florida) in June 2008 and January 2009. Indian River is a permanent saltwater site with salinities typically around 32 ppt. Adult L. goodei were collected from the Wakulla River at the Upper Bridge location (Wakulla County, Florida) in June 2008 and May 2010. This site is a freshwater river with a salinity of 0.2 ppt. All individuals were collected using dip nets and seines. Animals were transported back to the University of Illinois where they were housed by population in 38-L (10 gallon) and 110-L (29 gallon) aquaria. Fish were housed in their native salinity. Lucania goodei were maintained at 0 ppt, and L. parva were maintained at 35 ppt. In all experiments, our freshwater source was dechlorinated city water (water treated with Start Rite), and our saltwater source was reverse osmosis water from a four-stage barracuda RO/DI unit (Aqua Engineering and Equipment, Winter Park, FL) to which we added Instant Ocean® Sea Salt (Spectrum Brands, Atlanta, GA) to achieve the desired salinity. Salinity was verified with a YSI-63 salinity meter (YSI Inc., Yellow Springs, OH). Fish were fed ad libitum daily with frozen brine shrimp. Lights were maintained on a 14L:10D cycle.
FEMALE SPECIES RECOGNITION EXPERIMENT
In summer 2009, we performed no-choice mating trials using both conspecific and heterospecific crosses. “Cross” refers to the manner in which the two species were paired. We had four crosses: (1) L. goodei♀×L. goodei♂, (2) L. goodei♀×L. parva♂, (3) L. parva♀×L. goodei♂, and (4) L. parva♀×L. parva♂. Each cross was placed in either 0 or 15 ppt resulting in eight experimental treatments (four cross types × two salinity conditions). We used 15 ppt as our saltwater treatment because it is above the isosmotic point (10 ppt) but still within the range of salinities that adult L. goodei can tolerate (Kilby 1955; Fuller 2008b). Pairs of fish were put in 38-L aquaria (10 gallon) filled with the appropriate water treatment. All fish for this and the proceeding experiments were acclimated to their salinity treatment for at least 24 h. Previous research has shown that changes in osmoregulatory gene expression in a closely related species (Fundulus heterosclitus) occur within 24 h of salinity change (Scott et al. 2004a,b). Additionally, “plunge tests” where fish were rapidly transferred between salinities found that L. parva and L. goodei can tolerate rapid alterations in salinity within the limits used in this article (0–15 ppt) (Dunson and Travis 1991, R. Hale, pers. comm.).
We originally performed four replicates of each treatment resulting in 32 pairs of fish. We refer to these fish as “female species recognition—set 1.” These tanks were set up in May 2009, and tanks were checked for eggs every day. Due to unforeseen circumstances, we stopped collecting data after 14 days. During the hiatus, pairs remained together. At the end of June 2009, we resumed egg collection on “female species recognition—set 1” for another 41 days. We also set up additional four replicates (32 pairs of fish) of each treatment, which we refer to as “female species recognition—set 2” in late June 2009. We collected eggs from these tanks for 41 days.
From these data, we measured the rate of egg production and latency to mate. Egg production was calculated as the number of eggs collected from pairs of fish divided by the time span that the tank was actively monitored for eggs. For these analyses, we pooled the “female species recognition—set 1” and “female species recognition—set 2” trials resulting in 64 total pairs of fish. We also measured latency to mate as the number of days until eggs were first observed for the “female species recognition—set 2” trials. We excluded the “female species recognition—set 1” from this dataset due to the interruption in egg collection. If no eggs were present at the end of the 41-day experiment, then we conservatively assigned the trial a latency of 41 days. Females that did not lay any eggs during the experiment were included in both the latency to mate analyses and the egg production analyses as this lack of egg production likely reflects female mating preferences. Females were randomly assigned to treatments. Hence, variation in female breeding condition should not cause spurious treatment differences.
STATISTICAL ANALYSIS
These data were analyzed using generalized linear models in Proc Genmod in SAS version 9.2 (SAS institutes, Cary, NC). Both dependent variables had a truncated exponential distribution necessitating the use of the gamma distribution with a log link function. Analyses assuming a normal distribution produced qualitatively similar results. The model included the fixed effects of male species (L. parva, L. goodei), female species (L. parva, L. goodei), salinity (0 ppt, 15 ppt), and their interactions. Simple behavioral isolation predicts a significant interaction between male species and female species where egg production is high and latency to mate is low in conspecific crosses relative to heterospecific crosses. Environment-dependent behavioral isolation predicts a significant interaction between salinity, male species, and female species. All graphs shows means ± SEs.
MALE COMPETITION AND SPECIES RECOGNITION EXPERIMENT
We measured conspecific and heterospecific male competition as well as male species recognition. We established three male pair treatments: conspecific L. goodei (two L. goodei ♂s), conspecific L. parva (two L. parva ♂s), and a heterospecific pair (one L. goodei ♂ and one L. parva ♂). These pairs were placed in either fresh (0 ppt) or salt (15 ppt) water and observed over two days where they were exposed to a female L. goodei on one day and a female L. parva on the other. The order of female presentation was randomized. Because we were particularly interested in competition between males of the two species, we conducted twice as many heterospecific male pairs as we did of the conspecific pairs for each species. Each pair of males was tested with females of both species but was tested only in a single salinity.
Behavioral trials were conducted in 38-L aquaria (10 gallon). Fish were allowed to acclimate to these salinities at least 24 h before testing. Each trial was videotaped for 30 min using a Canon HG10 camcorder (Canon Inc., Tokyo, Japan). The next day, the procedure was repeated using a female of the alternate species. Some females were used in multiple trials (up to four trials for a single female in a single day). We noted whether females spawned. If a female failed to spawn in any of the trials, she was deemed unreceptive and not gravid and those observations were removed from the dataset. Aquaria were drained and refilled between male pairs to remove any chemical cues. Males were reused in the experiment but specific male pairs were never repeated. These experiments were performed in spring/summer of 2008, 2009, and 2010.
We planned to perform 10 replicates per salinity. However, due to difficulties in getting the females to spawn, we only ended up with 8.5 replicates in freshwater (eight full replicates and one trial where the L. goodei female spawned but the subsequent L. parva female did not). We had 10 full replicates for the saltwater treatment. Thus, we ended up with 148 trials (saltwater trials = 10 replicates × 2 female species × 4 male pairs = 80 trials; freshwater trials = 8.5 replicates × 2 female species × 4 male pairs = 68). We removed several trials where the camera had stopped recording before reaching 30 min (n= 5). We also removed several trials where the fish did not interact with each other in any way (n= 8). Overall, we removed 13 trials leaving us with 135 trials. After exclusions, there were 66 heterospecific trials and 69 conspecific trials (36 conspecific L goodei trials and 33 conspecific L. parva trials).
All videotapes were scored by E.B. using an event-recording program (JWatcher, http://www.jwatcher.ucla.edu/). We recorded male–male aggressive interactions (chasing, fin flares, sigmoid displays, and circle fights), courtship behaviors (head flicks and circle swims), and spawns (Foster 1967; McGhee et al. 2007; McGhee and Travis 2010; Supporting Information Text S1). From these data, we calculated the total number of aggressive behaviors (chasing + fin flares + sigmoid displays + circle fights) and the total number of courtship behaviors (head flicks + circle swims).
STATISTICAL ANALYSIS
We performed three separate analyses for (1) behavior in conspecific trials, (2) behavior in heterospecific trials, and (3) a combined analysis of conspecific and heterospecific behavior. For conspecific trials, we analyzed the sum total of all behavioral counts (i.e., total courtship and total aggression) between the two males in each trial. In the heterospecific trials, we could easily distinguish the two males, and we measured the amount of aggression, courtship, and spawning performed by each male in each trial. To compare the conspecific and heterospecific treatments, we analyzed the sum total of all behavioral counts for each trial.
AGGRESSION ANALYSIS
For conspecific male pairs, we analyzed the effects of male pair (conspecific L. goodei or conspecific L. parva), female species, salinity, and their interactions on total counts of aggression using a generalized linear model assuming a negative binomial error distribution with a log link function. The negative binomial model is appropriate for count data (Zuur 2009). A type 3 analysis was used to determine the significance of model terms. Similarly, we compared total counts of aggression among all three male pairs (conspecific L. goodei, conspecific L. parva, and heterospecific) using male pair, female species, salinity, and their interactions as fixed effects in our model assuming a negative binomial error distribution (see above).
For individual counts of aggression in heterospecific trials, we analyzed the effects of male species, female species, salinity, and their interactions on the amount of aggression displayed by males of each species using a repeated measures analysis. We used a repeated measures analysis because males who were attacked tended to counterattack. The covariance between males within a male pair was controlled using trial as a repeated factor in the analysis. Because this was also count data, we again used a generalized linear model that assumed a negative binomial distribution using SAS Proc Genmod. We specified “trial” as the repeated factor and analyzed the significance of our treatment effects using a type 3 analysis.
COURTSHIP ANALYSIS
The courtship data for both contexts was heavily weighted with zeros. To analyze these data, we used a hurdle model with a negative binomial distribution in R (R Development Core Team 2009). In this analysis, a binomial model with a logit link function is used to measure the probability of getting a zero, and a count process is used to model the nonzero values (Zuur 2009). The nonzero values are modeled using a truncated negative binomial distribution with a log link. We initially used a binomial model that included male pair, female species, salinity, and their interactions. However, using AIC criteria, we found that dropping salinity and its interactions from the model improved model fit. The results from the full model were qualitatively identical to the model that excluded salinity and its interactions. The final binomial model for our conspecific data contained the following fixed effects as predictors of zeros: male pair, female species, and the interaction between the two. For the count process, male pair, female species, salinity, and their interactions were modeled as fixed effects. For the count model of courtship in conspecific pairs, the nonsignificant three-way interaction (male pair × female species × salinity) had to be removed in order for the model to converge.
Similar models were used for the heterospecific data except that male species was used in place of male pair. As with the conspecific data, the binomial model had male species, female species, and their interactions as predictors of zeros. Including salinity in the binomial model did not alter the qualitative results, but decreased model fit. The count model used male species, female species, salinity, and all the interactions as main effects.
Finally, we examined the effects of date in all our models. It was not statistically significant and had no appreciable effects on our analyses, so we removed it. The raw data for these experiments have been deposited at Dryad (Dryad doi:10.561/dryad.fq613m13).
STRENGTH OF BEHAVIORAL ISOLATION
We calculated total behavioral isolation using the IPSI statistic (for equations see Rolan-Alvarez and Caballero 2000). IPSI has been shown to be one of the most unbiased statistics for measuring behavioral isolation especially when small sample sizes are used (Perez-Figueroa et al. 2005). We used JMATING software (Carvajal-Rodriguez and Rolan-Alvarez 2006) and 10,000 rounds of bootstrap resampling to estimate the mean and SD of behavioral isolation. The IPSI statistic ranges from –1 (full disassortative mating) to 0 (random mating) to 1 (full assortative mating). The program considers the number of potential matings between males and females of each species in comparison to the realized number of matings. We used the data from our male competition data. For conspecific trials (where two conspecific males were placed in an aquarium with either a conspecific or heterospecific female), we considered there to be one potential mating opportunity because it was impossible to distinguish between the two males. For the heterospecific aggression trials (where one L. parva and one L. goodei male were placed with either an L. parva or an L. goodei female), we considered there to be two potential mating opportunities: the female could mate with either a conspecific or a heterospecific male. We calculated IPSI three times: once including solely trials conducted in freshwater, once including solely trials conducted in saltwater, and once including all trials regardless of salinity. We used a G-test of independence to determine if the IPSI values for freshwater and saltwater were significantly different (Sokal and Rohlf 1981). The G-test was conducted using the same tables of realized mating opportunities used to estimate IPSI.
COMPARISON OF DIFFERENT REPRODUCTIVE ISOLATING BARRIERS
We quantified several reproductive isolating barriers using stage-specific indices of reproductive isolation. These indices vary from –1 to 0 to 1 with 0 representing no barrier to gene flow and 1 representing a complete barrier. We then calculated the strength and absolute contribution of each reproductive isolating barrier to overall reproductive isolation (Ramsey et al. 2003). Full details on our calculations of reproductive isolation can be found in the Supporting Information (Text S2).
We used the strength of each reproductive barrier to estimate the cumulative reproductive isolation that has evolved in this system. Following equations (1–6) in Ramsey et al. (2003), the absolute contribution of each reproductive barrier was calculated, taking into account the timing of its contribution in the life cycle. Barriers that occur earlier in the life cycle make a larger contribution to total reproductive isolation. Conversely, barriers occurring later in the life cycle make a small contribution to total reproductive isolation simply because there is less potential hybridization/introgression for them to prevent. We assumed that our barriers fall in the following order: geographic isolation, behavioral isolation, intrinsic isolation.
Results
FEMALE SPECIES RECOGNITION
Both indices of female species recognition (egg production and latency to mate) showed strong and symmetrical behavioral isolation (Table 1; Fig. 1A, B). Females of both species produced significantly more eggs (and had a lower latency to mate) with conspecific males than with heterospecific males. The interaction between male species and female species was highly significant for both indices. The fact that the male species × female species × salinity term was nonsignificant for both indices indicates that the strength of behavioral isolation was not influenced by the external salinity.
| Egg production | Latency to mate | ||||
|---|---|---|---|---|---|
| Parameter | df | χ2 | P | χ2 | P |
| Male species | 1 | 9.06 | 0.0026 | 0.01 | 0.9404 |
| Salinity | 1 | 3.97 | 0.0465 | 3.74 | 0.0533 |
| Male × salinity | 1 | 1.02 | 0.3126 | 1.29 | 0.2565 |
| Female species | 1 | 2.08 | 0.1493 | 3.45 | 0.0633 |
| Male species ×female species | 1 | 7.44 | 0.0064 | 9.85 | 0.0017 |
| Salinity × female species | 1 | 4.04 | 0.0444 | 4.88 | 0.0272 |
| Male species ×salinity × female species | 1 | 0.56 | 0.4525 | 1.40 | 0.2372 |
- Note: Significant parameters are indicated in bold.
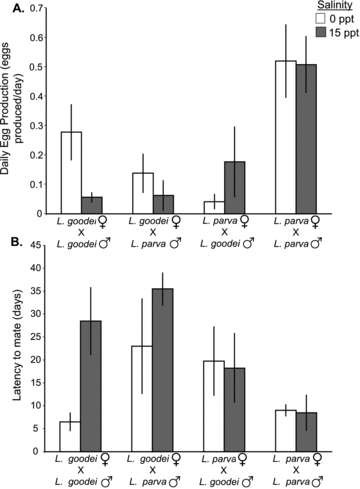
Indices of female species recognition from the no-choice experiment. (A) Average daily egg production. (B) Latency to mate in days. N= 8 for all treatment combinations. Error bars ± SE.
Salinity and an interaction between female species and salinity affected both latency to mate and egg production (Table 1). These effects were driven by the fact that L. goodei females produced fewer eggs (and had a greater latency to mate) in salt water than in fresh water (Fig. 1A, B). In contrast, L. parva females produced similar amounts of eggs (and had a similar latency to mate) in both salinity treatments. Egg production of conspecific pairs at 0 ppt was within the normal range for these species (Arndt 1971; E. L. Berdan, pers. obs.). There were also significant main effects of both male species and salinity on egg production. Male L. parva induced females to lay more eggs overall than L. goodei males.
MALE–MALE AGGRESSION
Male conspecific aggression was affected by the interaction between male species and female species. The pattern generally supported the idea of behavioral isolation where males compete more intensely for females of their own species. Rates of aggression in the conspecific male pairs differed between species and were influenced by the species of the female and the salinity (Fig 2A; Table 2). Overall, L. parva males were more aggressive with each other than were L. goodei males. Lucania parva males were 1.7 times more aggressive in the presence of L. parva females than in the presence of L. goodei females, but the result did not reach statistical significance (χ21= 2.88, P= 0.0899, see Table S1). In contrast, L. goodei were 6.2 times more aggressive in the presence of L. goodei females than in the presence of L. parva females, and the result was highly significant (χ21= 22.09, P= 0.0001). Salinity also had different effects on aggression in the two species where L. parva was more aggressive in 15 ppt than in 0 ppt (χ21= 3.9, P= 0.0484), and L. goodei was more aggressive in 0 ppt than in 15 ppt (χ21= 3.97, P= 0.0462).
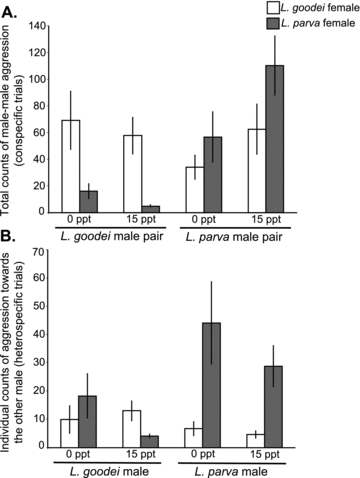
Aggression in conspecific and heterospecific trials. (A) Total aggression in conspecific trials. Sample sizes are as follows (LG =L. goodei, LP =L. parva): LG ♂ pair–LG ♀–0 ppt = 9, LG ♂ pair–LG ♀–15 ppt = 10, LG ♂ pair–LP ♀–0 ppt = 8, LG ♂ pair–LP ♀–15 ppt = 9, LP ♂ pair–LG ♀–0 ppt = 7, LP ♂ pair–LG ♀–15 ppt = 9, LP ♂ pair–LP ♀–0 ppt = 8, LP ♂ pair–LP ♀–15 ppt = 9. (B) Aggression per male in heterospecific trials. Sample sizes are as follows: LG ♂–LG ♀–0 ppt = 16, LG ♂–LG ♀–15 ppt = 17, LG ♂–LP ♀–0 ppt = 16, LG ♂–LP ♀–15 ppt = 17, LP ♂–LG ♀–0 ppt = 16, LP ♂–LG ♀–15 ppt = 17, LP ♂–LP ♀–0 ppt = 16, LP ♂–LP ♀–15 ppt = 17. Error bars ± SE.
| Parameter | df | χ2 | P |
|---|---|---|---|
| Male pair | 1 | 15.01 | 0.0001 |
| Female species | 1 | 9.05 | 0.00026 |
| Male pair × female species | 1 | 23.85 | <0.0001 |
| Salinity | 1 | 0.02 | 0.8912 |
| Male pair × salinity | 1 | 7.82 | 0.0052 |
| Female species × salinity | 1 | 0.2895 | 0.2895 |
| Male pair × female species × salinity | 1 | 1.41 | 0.2356 |
- Note: Significant parameters are indicated in bold.
In heterospecific interactions, males of both species were more aggressive in the presence of conspecific females. Figure 2B shows the average level of aggression for each species in each salinity and in the presence of each type of female. Overall aggression in L. parva males was higher in the presence of conspecifics with Lucania parva males dominating over L. goodei males in the presence of L. parva females (see also Table 3; Fig. 3). The pattern was more complex with L. goodei. Figure 2B shows that overall male L. goodei displayed more aggression in the presence of L. parva females in 0 ppt. However, an examination of the differences in aggression (i.e., which male was more aggressive; Fig. 3) shows that L. goodei males were more likely to dominate L. parva males in the presence of L. goodei females. The heightened aggression of L. goodei in the presence of L. parva females (Fig. 2B) is most likely a response to the high levels of aggression from L. parva males when L. parva females are present (i.e., fish that are attacked tend to counterattack). Figure 3 also shows little effect of salinity on the outcome of heterospecific aggression.
| Parameter | df | χ2 | P |
|---|---|---|---|
| Male species | 1 | 4.2 | 0.0403 |
| Female species | 1 | 4.87 | 0.0273 |
| Male species × female species | 1 | 13.99 | 0.0002 |
| Salinity | 1 | 1.88 | 0.1707 |
| Male species × salinity | 1 | 0.41 | 0.5209 |
| Female species × salinity | 1 | 1.88 | 0.1705 |
| Male species × female species × salinity | 1 | 5.18 | 0.0229 |
- Note: Significant parameters are indicated in bold.

Raw differences in aggression in heterospecific trials (no. of L. goodei aggressive behaviors – no. of L. parva aggressive behaviors) as a function of salinity and female species. Each dot is a single trial.
When comparing all three types of male pairs (conspecific L. parva, conspecific L. goodei, and heterospecific), conspecific L. parva male pairs had the highest total levels of aggression, conspecific L. goodei male pairs were intermediate, and heterospecific pairs had the lowest levels of aggression (Fig. 4; Table S2, male pair: χ21= 14.37, P= 0.0008). Again, aggression was highest in conspecific male pairs tested with conspecific females (Fig. 4; Table S2, male pair × female species: χ21= 30.75, P= 0.0001). Although aggression tended to vary as a function of male pair and salinity (χ21= 5.74, P= 0.0566), there was no significant interaction between male pair, female, and salinity (χ21= 1.02, P= 0.6).
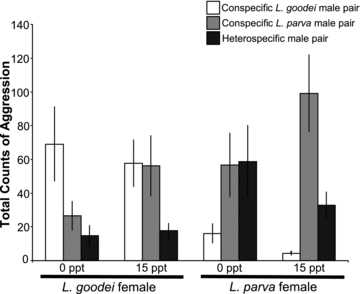
Total counts of male–male aggression across conspecific and heterospecific trials. Error bars ± SE.
COURTSHIP
Lucania goodei males actively courted L. goodei females but did not court L. parva females (Fig. 5A, B; Tables 4 and 5). In contrast, L. parva males courted both types of females. Patterns of courtship were identical in both the conspecific and the heterospecific context. Although salinity and its interactions were not significant for any courtship dataset, graphical examination of the data suggested that L. parva males may be more discriminating between females at 15 ppt than at 0 ppt. To test this, we conducted a post-hoc two-tailed t-test. In the conspecific context, L. parva males were more likely to discriminate between the two species at 15 ppt (post-hoc two-tailed t-test assuming unequal variances: 0 ppt—T= 0.671, df = 13, P= 0.514; 15 ppt—T= 2.66, df = 10, P= 0.024). The same was true in the heterospecific context (post-hoc two-tailed t-test assuming unequal variances: 0 ppt—T= 1.56, df = 21, P= 0.133; 15 ppt—T= 3.62, df = 25, P= 0.001). However, analyses restricted solely to L. parva found no statistically significant interaction between female and salinity (salinity × female interaction heterospecific data; χ21= 0.63, P= 0.426, salinity × female interaction conspecific data; χ21= 1.54 P= 0.215). A power analysis on the heterospecific data indicated that sample sizes would need to be increased fourfold (more than 250 trials and 500 males) to detect a significant male species × female species × salinity interaction. For the conspecific data, increasing sample sizes 20-fold (more than 1300 male pairs) would still not result in sufficient power to detect a significant male species × female species × salinity interaction.
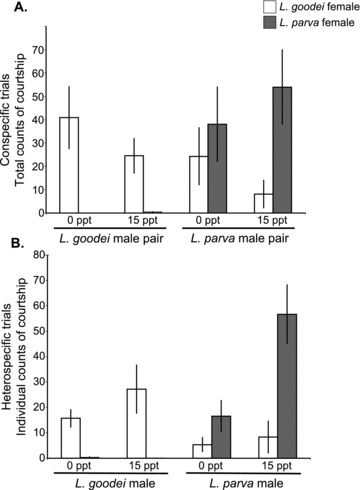
Courtship in conspecific and heterospecific trials. (A) Courtship totals in conspecific trials. Sample sizes are the same as in (2A). (B) Courtship per male in heterospecific trials. Sample sizes are the same as in (2B). Error bars ± SE.
| A. Count Model (truncated negative binomial distribution with log link) | |||
|---|---|---|---|
| Parameter | df | Z | P |
| Male pair | 1 | −0.513 | 0.608 |
| Female species | 1 | −2.759 | 0.0058 |
| Male pair×female species | 1 | 3.252 | 0.0012 |
| Salinity | 1 | −0.788 | 0.431 |
| Male pair×salinity | 1 | 0.076 | 0.938 |
| Female species×salinity | 1 | 0.60 | 0.546 |
| B. Zero hurdle Model (binomial distribution with logit link) | |||
|---|---|---|---|
| Parameter | df | Z | P |
| Male pair | 1 | −2.083 | 0.037 |
| Female species | 1 | −3.76 | 0.0002 |
| Male pair×female species | 1 | 4.284 | <0.0001 |
- Note: Significant parameters are indicated in bold.
| A. Count Model (truncated negative binomial distribution with log link) | |||
|---|---|---|---|
| Parameter | df | Z | P |
| Male species | 1 | −0.204 | 0.838 |
| Female species | 1 | −1.064 | 0.287 |
| Male species×female species | 1 | 1.15 | 0.250 |
| Salinity | 1 | 1.133 | 0.257 |
| Male species×salinity | 1 | −0.186 | 0.852 |
| Female species×salinity | 1 | −0.07 | 0.946 |
| Male species×female species×salinity | 1 | 0.071 | 0.943 |
| B. Zero hurdle Model (binomial distribution with logit link) | |||
|---|---|---|---|
| Parameter | df | Z | P |
| Male species | 1 | −3.961 | <0.0001 |
| Female species | 1 | −5.093 | <0.0001 |
| Male species×female species | 1 | 6.443 | <0.0001 |
- Note: Significant parameters are indicated in bold.
STRENGTH OF REPRODUCTIVE ISOLATION
Behavioral isolation (IPSI) was high. In fresh water, IPSI (± 1 SD) was 0.87 ± 0.14. In saltwater, IPSI was 0.93 ± 0.07. Across both salinities, IPSI was 0.91 ± 0.07. Estimates of behavioral isolation were not significantly different between freshwater and saltwater (G= 0.325, df = 3, P= 0.955). Sympatric populations may occur in fresh or brackish water so we used our final IPSI value (encompassing both salinities) to estimate behavioral isolation for the Ramsey et al. (2003) calculations.
Geographic isolation was also high. In Florida, geographic isolation was 0.91. Overall intrinsic isolation was substantial (0.66), although it was lower than both behavioral and geographic isolation. Reduced hybrid mating success and reduced fitness of back-cross and F2 offspring made the largest contributions to intrinsic isolation (F1 larval survival = 0.06, F1 survival to adulthood =–0.07, F1 mating success = 0.38, F1 offspring survival = 0.28; see Table S3). The total strength of reproductive isolating barriers is 0.997 (Table 6).
| Barrier | Strength | Absolute contribution |
|---|---|---|
| Geographic | 0.91 | 0.911 |
| Behavioral | 0.91 | 0.081 |
| Intrinsic | 0.66 | 0.005 |
| Total RI | 0.997 |
Discussion
Our study demonstrated that (1) behavioral isolation and overall reproductive isolation are strong in this system, (2) male species recognition strongly contributes to behavioral isolation, (3) male species recognition is very asymmetrical, and (4) behavioral isolation did not vary with salinity. These results suggest that adaptation to salinity is not directly related to the development of behavioral isolation in this system. Instead, our results suggest that male preference for conspecific females may have evolved in part via reinforcement due to the low fitness of hybrids produced by L. goodei males and L. parva females.
SALINITY, ADAPTATION, AND REPRODUCTIVE ISOLATION
Reproductive isolation between L. goodei and L. parva is nearly complete (Total RI = 0.997). Behavioral isolation is very high in Lucania (0.91). To compare the level of behavioral isolation in Lucania to other systems, we conducted a literature survey of other recently published IPSI values (Table S4). When compared with other IPSI values, behavioral isolation in Lucania was stronger than 87% of other recently published between species comparisons (see Table S4). In Lucania, behavioral isolation is approximately 1.5× stronger than intrinsic isolation (Table 6). Hence, this study joins the many others showing that behavioral isolation is higher than postzygotic isolation between closely related species (Mendelson 2003; Stelkens et al. 2010). However, we also note that postzygotic isolation is substantial in Lucania and that intrinsic isolation mostly resulted from decreased F1 hybrid mating success and reduced back-cross and F2 hybrid survival. Studies that only consider intrinsic isolation in the early developmental stages of F1 offspring frequently find low estimates of intrinsic isolation (Wiley et al. 2009). Of course, we cannot determine the order in which these isolating barriers arose and which contributed most heavily to the initial stages of divergence.
We used wild-caught animals for this experiment and, thus, cannot determine whether behavioral isolation is due to genetic effects or due to environmental effects such as learning. In the speciation literature, learning has usually been discussed as an alternative hypothesis to reinforcement for explaining heightened behavioral isolation in sympatry. Under this scenario, animals in sympatry learn to avoid mating with heterospecifics. In the current study, we used animals from allopatric populations that had no opportunity to learn to discriminate between the two species. Alternatively, learning could still influence the results of this study if individuals learn to prefer similar killifish that they encounter in their populations. Under this alternative scenario, learning leads to lower behavioral isolation in sympatry where fish develop while experiencing heterospecific Lucania. There is no evidence for this in our system. Fuller et al. (2007) found no difference in behavioral isolation between allopatric and sympatric L. goodei/L. parva population pairs even though fish from sympatric populations had experienced heterospecifics and were housed with heterospecifics. Additionally, a recent study using “highly allopatric” populations (e.g., fish must swim >160 km to encounter heterospecific Lucania) found that behavioral isolation is lower in extremely allopatric species pairs than in sympatric species pairs (which is consistent with reinforcement, see discussion below, O. Gregorio, unpubl. data). Although we cannot fully exclude learning, it is unlikely to play a large role in behavioral isolation.
Salinity affected both male and female behaviors but had no appreciable effects on behavioral isolation. Lucania goodei females spawned fewer eggs and had a higher latency to mate at 15 ppt than at 0 ppt regardless of the species identity of the male. Both L. goodei and L. parva males were more likely to engage in conspecific aggression when placed in the salinity most similar to the site from where they were collected (0 ppt for L. goodei, 15 ppt for L. parva). Previous work in Lucania indicates local adaptation where L. goodei has higher survival in fresh water than L. parva and vice versa in salt water (Fuller et al. 2007; Fuller 2008a,b; Kozak et al. 2012). Our current study extends the effects of salinity to female fecundity and behavior. Osmoregulation is critical for organism homeostasis and being placed at a nonoptimal salinity is costly (Grizzle and Altinok 2003) particularly for L. goodei.
Although salinity affected multiple behaviors, these did not affect behavioral isolation. Environment-dependent behavioral isolation should have resulted in three-way interactions between male species, female species, and salinity for variables directly relevant to behavioral isolation. We tested three-way interactions for the egg production, latency to mate, and male courtship measures. We also examined male courtship for each species singly and found no interaction between salinity and female species for either L. goodei or L. parva courtship levels (in both the conspecific and heterospecific contexts). These results suggest that salinity has little effect on behavioral isolation. The one exception was that L. parva males tended to discriminate more between conspecific and heterospecific females at 15 ppt than at 0 ppt. Whether this is due to salinity altering signaling dynamics is unclear. However, this result should be interpreted with caution. t-tests showed statistically significant difference in male L. parva courtship at 15 ppt but not at 0 ppt in both the conspecific and heterospecific assays. Again, the interaction term in our model (male species × female species × salinity) was nonsignificant. A power analysis indicated that a very large sample size was needed to detect this interaction. Thus even if this trend is real, it is weak and unlikely to be biologically significant.
There are a number of ways for ecological selection to drive behavioral isolation that do not result in environment dependent behavioral isolation. Under a good genes for ecologically relevant traits model, the expression of female preferences may rely on male traits that are plastic. The expression of traits and preferences would then be sensitive to the salinity experienced in early development in which case our experiment would be unable to detect environmental-dependent behavioral isolation because we used wild-caught animals. Although this scenario is possible, we think it unlikely. We have measured preferences of L. parva from both freshwater and saltwater populations and found robust behavioral isolation in both cases (Fuller et al. 2007; G.M Kozak pers. comm.). However, different environmental conditions may also result in genetic differentiation of traits and preferences that are not plastic in expression (Merrill et al. 2011). This would make it impossible to pick up the signature of good genes for ecologically relevant traits with this experiment. Another possibility is that ecological selection has occurred as a function of another environmental variable. Obviously, we cannot definitively rule out the possibility that behavioral isolation is driven by ecological selection. However, our study provides little evidence for the hypothesis that behavioral isolation was driven by differential adaptation to salinity via a sensory drive model. If behavioral isolation has evolved from a nonecological mechanism, then another mechanism must be instrumental in bringing the genes involved in adaptation and reproductive isolation into linkage disequilibrium.
ASYMMETRICAL MALE SPECIES RECOGNITION
We found robust male species recognition that was strongly asymmetrical. Lucania parva males preferred conspecific females, but readily courted L. goodei females. However, L. goodei males failed to court L. parva females. The direction of this asymmetry in male species recognition is concordant with (i.e., it matches) the direction of an asymmetry in postzygotic isolation (Fuller 2008a). Hybrid males produced from crosses between an L. goodei male and an L. parva female have greatly decreased fertility compared to males of the reverse hybrid cross (L. parva♂×L. goodei♀) as well as males of the two parental species (Fuller et al. 2007; Fuller 2008a). Reinforcement may have created this pattern due to asymmetrical intrinsic isolation.
The evolution of behavioral isolation via reinforcement is a nonecological mechanism. Of course, the postzygotic isolation driving this process may have arisen due to divergent, ecological selection as a function of different environmental conditions. Although extrinsic isolation is present in Lucania, it cannot account for the asymmetry in behavioral isolation. The survival of both F1 crosses is very high and is robust to external salinity. Hybrids carrying a large proportion of the L. goodei genome suffer reduced fitness in saltwater, but this does not predict the asymmetry shown here.
Reinforcement can only occur when there is gene flow and hybridization between two species/populations. Thus, allopatric populations such as ours should lack the signature of reinforcement. We chose these allopatric populations for this study because we wanted to examine populations that were adapted to very different environmental conditions. Although neither of our source populations is sympatric, reinforcement may be occurring in our allopatric populations in several ways. First, both populations may be undergoing reinforcement caused by the occasional immigration of migrants from close, heterospecific populations (see Fuller and Noa 2008 for a map showing heterospecific populations within 4 km). Low levels of migration have been shown to increase the chances of reinforcement (Felsenstein 1981; Kelly and Noor 1996; Servedio and Kirkpatrick 1997). Alternatively, reinforcement could be occurring only in sympatric sites with alleles for species recognition spreading to nearby allopatric sites (Walker 1974; Hoskin et al. 2005). A final possibility is that our allopatric populations were colonized by fish from sympatric populations as sea-levels retreated during the Quaternary (Burgess and Franz 1978).
Demonstrating reinforcement is a challenging task and requires that (1) hybridization and gene flow occur in sympatric populations, (2) hybrids have reduced fitness, (3) prezygotic isolation is increased in areas of sympatry, (4) variation in preference is heritable, and (5) displacement has not occurred for other reasons such as ecological gradients (see Howard 1993; Servedio and Noor 2003; Coyne and Orr 2004). The biology of L. goodei and L. parva meets many of these requirements. Heterospecific matings (and gene flow) occur in natural populations (Hubbs et al. 1943; R. C. Fuller, unpubl. data). Hybrids have decreased fitness (Fuller 2008a). Female mating preferences for conspecifics are heritable (R. C. Fuller, unpubl. data), although whether the same is true for males is unknown. The critical issue is whether prezygotic isolation is heightened in areas of sympatry. We are currently performing experiments to test this and are conducting genetic crosses to further examine the genetic basis of species recognition by both males and females. Preliminary evidence suggests that “highly allopatric” population pairs where fish must swim over fifty kilometers to experience a heterospecific have lower levels of behavioral isolation than sympatric populations or allopatric populations that are in close proximity to sympatric populations.
SPECIES RECOGNITION IN THE LUCANIA SYSTEM
Our study demonstrated high levels of male mating preference for conspecific females that contribute to behavioral isolation. Traditionally females are assumed to be the “choosy sex,” and most work on reinforcement and sexual selection focuses on female choice/species recognition (Andersson 1994; Ord and Stamps 2009). However, there is a growing literature documenting male choice (Engqvist and Sauer 2001; Wedell et al. 2002; Wong and Jennions 2003; Byrne and Rice 2006; Espinedo et al. 2010) as well as reinforcement of male mating preferences (Peterson et al. 2005; Servedio 2007; Svensson et al. 2007). A recent meta-analysis showed that male discrimination of heterospecifics (rather than female discrimination) was more common in closely related species than in more distantly related species (Ord et al. 2011). A cost to male courtship/mating (e.g., sperm/ejaculate costs, exposure to predators, time lost) is often needed for male mate choice/species recognition to evolve (Kokko and Johnstone 2002; Byrne and Rice 2006). Although, male species recognition has most likely evolved via reinforcement, it is unclear how mating with heterospecifics reduces male fitness. Lucania males guard breeding territories but provide little (if any) care to the offspring. This suggests that mating with heterospecifics either entails other costs such as lost mating opportunities with conspecifics or sperm depletion that affects fertilization success.
Finally, our two experiments gave different pictures of behavioral isolation. The female species recognition experiment showed that behavioral isolation was both symmetrical and robust to environmental changes. In contrast, the male competition experiment indicated that behavioral isolation was strongly asymmetric with L. goodei males refusing to court L. parva females. The differing results of our two experiments are most likely due to two factors: time frame and experimental set-up. Our female species recognition experiment consisted of a no-choice test that ran for 41 days. In contrast, individual trials in our male competition experiment lasted for 30 min. No-choice mating assays are conservative measures of behavioral isolation because individuals are forced to choose whether to mate (Houde 1997; Coyne and Orr 2004). In nature, male/female interactions are usually brief, and other potential mates are in close proximity (Arndt 1971; Fuller 2001). Hence, the male competition experiment, with its 30-min observation period, may give a more accurate picture of behavioral isolation. Additionally, our male competition experiment included direct observation of the fish that led to the discovery that male species recognition is an important component of behavioral isolation in this system.
CONCLUSIONS
We found high levels of behavioral isolation between L. goodei and L. parva, but behavioral isolation did not vary appreciably with salinity. Male species recognition (i.e., the choice of whether to court a female) played a large role in behavioral isolation and was asymmetric. Lucania goodei males did not court L. parva females, but L. parva males did court L. goodei females. This asymmetric behavioral isolation mirrors the pattern of intrinsic postzygotic isolation observed by Fuller (2008a) who found lower fitness of offspring from hybrid crosses between male L. goodei and female L. parva than in the reciprocal hybrid cross. Hence, asymmetric postzygotic isolation may have resulted in asymmetric behavioral isolation due to reinforcement.
Associate Editor: C. Klingenberg
ACKNOWLEDGMENTS
Thanks to A. Bell, A. Johnson, G. Kozak, M. Schrader, D. Welsh, M. Zhou, and three anonymous reviewers for helpful comments that improved the manuscript. This work was approved by the University of Illinois IACUC (No. 08183). This work was funded by the University of Illinois and two National Science Foundation Awards (DEB 0953716 and DEB 1110658).




