Higher Capillary Electrophoresis Injection Settings as an Efficient Approach to Increase the Sensitivity of STR Typing
Abstract
Abstract: Evidentiary traces may contain low quantities of DNA, and regularly incomplete short tandem repeat (STR) profiles are obtained. In this study, higher capillary electrophoresis injection settings were used to efficiently improve incomplete STR profiles generated from low-level DNA samples under standard polymerase chain reaction (PCR) conditions. The method involves capillary electrophoresis with higher injection voltage and extended injection time. STR peak heights increased six-fold. Inherent to the analysis of low-level DNA samples, we observed stochastic amplification artifacts, mainly in the form of allele dropout and heterozygous peak imbalance. Increased stutter ratios and allele drop-in were rarely seen. Upon STR typing of 10:1 admixed samples, the profile of the major component did not become overloaded when using higher injection settings as was observed upon elevated cycling. Thereby an improved profile of the minor component was obtained. For low-level DNA casework samples, we adhere to independent replication of the PCR amplification and boosted capillary electrophoresis.
In forensic disputes, the evidentiary items can hold bodily fluids or contact traces. Especially from handled items, the DNA recovery is often low, resulting in incomplete DNA profiles in standard forensic procedures. Several options exist to enhance the sensitivity of DNA genotyping. First, the extract containing the DNA can be concentrated (1) by various methods after which fewer polymerase chain reaction (PCR) amplifications are possible, a disadvantage that can be met, in part, by using reduced-volume reactions (2,3). Second, the PCR method can be adjusted, and the techniques vary from nested PCR and whole genome amplification (4,5) to the use of an increased number of amplification cycles (6–8). Third, post-PCR purification has been put forward as a simplified approach for low-level DNA analysis (9), and in our laboratory ChargeSwitch magnetic beads (Invitrogen, Breda, The Netherlands) were successfully used to increase peak heights (PH) three-fold (C. C. G. Benschop and T. Sijen, unpublished results). The most popular method is to elevate the number of PCR cycles from 28 to 34, which is based on the observation that 34 cycles are sufficient to analyze the DNA present in a single diploid cell (6). The above-described methods effectively improve DNA profiling, but artifacts, such as allele dropout, heterozygous peak imbalance, increased stutter peaks, and sporadic contamination (allele drop-in) (6–8,10) are observed. These issues are generally dealt with by generating a so-called consensus profile based on multiple independent PCRs (11).
In this study, we explored the effects of using higher electrokinetic injection settings to increase the sensitivity of genotyping (12). Short tandem repeat (STR) PH would increase with minimal handling, costs, and consumption of PCR product. We compared characteristics of low-level DNA genotyping data obtained using boosted injection and more amplification cycles. We examined the possibilities of improving the genotyping data of the minor component in 10:1 admixed samples (13–15).
Materials and Methods
DNA Samples and STR profiling
Three pristine DNAs were used: DNA 007, the reference DNA in the AmpFlSTR®SGM Plus™ (SGM+) kit (Applied Biosystems [AB], Nieuwerkerk aan de IJssel, The Netherlands), human DNA (hDNA), the reference DNA in the Quantifiler™ kit (AB), and DNA9947A representing the reference DNA in the AmpFlSTR®Profiler™ (Profiler) kit (AB). Mock casework samples consisted of plastic straws and cups, from which volunteers (with known STR profile) drank once, and of plastic tie wraps, which the volunteers pulled once fiercely. For negative controls unused items were sampled. All items were UV irradiated for 3 h prior to usage. This procedure did not remove all DNA contamination (F. Beemster and T. Sijen, unpublished results). Samples were taken using standard cotton swabs and isolated with the QiaAmp 96 DNA Swab Biorobot Kit on a Qiagen Biorobot Universal System (Qiagen, Venlo, The Netherlands).
DNA quantification of DNA isolations was performed using the Quantifiler kit (AB), and analysis was performed on a 7900 real time PCR (AB). STR profiling was performed using the SGM+ kit (AB). For 28 + 6 amplifications, 10 μL of 28-cycling PCR product was transferred to a new tube, 0.5 μL of fresh AmpliTaq polymerase (AB) was added, and the SGM+ PCR protocol was applied for six cycles.
Performa DTR gel filtration cartridges (Edge BioSystems, Gaithersburg, MD) were used to remove residual dye molecules that cause the presence of dye-blobs. To remove all storage liquid, the prehydrated cartridges were centrifuged for 3 min at 1600 × g, placed on a new microtube, centrifuged for 2 min at 1600 × g, and transferred to a collection tube. An aliquot of 5 μL PCR product was added to the cartridge and collected by centrifugation for 2 min at 850 × g.
Capillary Electrophoresis
Electrophoresis was performed on a 3130XL genetic analyzer (AB). Injection settings varied as described in the experiments. Injection mixtures for standard 3 kV, 10 sec injections consisted of 1 μL PCR product, 0.4 μL Gene-Scan™-500 size marker (AB), and 8.6 μL HiDi-formamide (AB). For 9 kV/15 sec injections, 1 μL PCR product, 0.05 μL Gene-Scan™-500, and 8.95 μL HiDi-formamide were mixed. For 9 kV/15 sec injections of Performa DTR gel filtration cartridges purified PCR mixtures, 2 μL product, 0.02 μL Gene-Scan™-500, and 7.98 μL HiDi-formamide were mixed. The number of allelic ladder in runs with higher injection settings or purified PCR products was reduced 20-fold. Samples were denatured for 4 min at 98°C and rapid-cooled on ice blocks.
STR Typing Analysis
GeneMapper ID® Version 3.2.1 software (AB) was used to analyze STR profiles and determine the PH. Heterozygote balance, calculated as lowest peak/highest peak, was determined when both concordant alleles in heterozygous loci were present.
All STR profiles that were based on 28 cycling PCR products were analyzed using the marker specific stutter ratios provided by AB GeneMapper ID® software (D3S1358, 11%; vWA, 11%; D16S539, 13%; D2S1338, 15%; amelogenin (AMEL), 0%; D8S119, 12%; D21S11, 13%; D18S51, 16%; D19S433, 17%; THO1, 6%; FGA, 11%). In profiles based on 34 or 28 + 6 cycling conditions, stutter percentages were 1.5 times of these marker-specific ratios.
Our casework interpretation guidelines aimed to prevent false homozygote calling by removing allele calls for all single peaks below 100 rfu and single peaks between 100 and 175 rfu when uncalled peaks were visible. These interpretation guidelines were applied to the mock casework samples. In profiles obtained by boosted injection or elevated cycling, all peaks above the allele calling threshold of 50 rfu were included in the genotyping result. To enable direct comparison of the sensitivity of allele detection in amplifications of pristine DNA samples and also for the STR profiles obtained by standard procedure (28 cycles, 3 kV/10 sec injection), all alleles above 50 rfu were included in the genotyping data.
Baseline noise was determined as the maximum rfu value for the FAM, JOE, and NED channels in the region 250 to 295 nt of electropherograms of negative amplifications. This region was chosen because it is uninfluenced by dye-blobs or size marker pull-up peaks.
When determining the percentage of concordant alleles present in a SGM+ STR profile, homozygous alleles were counted as 2. Locus dropout was calculated as 0 alleles detected for that locus. A dropout allele refers to an undetected allele in a heterozygous locus where the other allele was called. Drop-in alleles refer detected nonconcordant alleles. Drop-in alleles could occur due to polymerase slippage (such as stutters at −1 position that were 4 nt shorter PCR products or stutters at +1 position that were 4 nt longer PCR products), or due to sporadic DNA contamination of sampled items.
A consensus profile in our laboratory consisted of alleles called upon standard STR profiling and alleles detected in n-1 of the profiles obtained after high sensitivity analysis in which n stands for the number of PCR repetitions performed.
Results and Discussion
Higher Injection Settings for Capillary Electrophoresis
For an ABI 3130xl genetic analyzer (AB), recommended injection settings resided between 1–3 kV and 3–22 sec, and the standard injection setting in our laboratory was 3 kV/10 sec. However, the apparatus allowed injections up to 15 kV for 600 sec. Injection conditions affect peak shape; generally higher injection voltages sharpen peaks while longer injection times broaden peaks. We raised the injection voltage from 3 to 6, 9, 12, and 15 kV and the injection time from 10 to 15, 20, 25, 30, 40, 50, 60, 120, and 300 sec, and found that injection at 9 kV during 15 sec resulted in peak shapes that still allowed correct binning and discrimination from background structures like spikes and blobs, while improving sensitivity (Fig. 1A and B). Boosted injection did not result in an increase in baseline noise nor in the presence of contaminating alleles in 30 negative amplification controls. Therefore, the detection threshold for standard STR analysis, which was at 50 rfu in our laboratory, was not changed. Dye-blobs that were present in profiles obtained using standard settings intensified upon boosted injection. They occurred at predictable positions, and could be discriminated from true alleles by peak shape. The residual dye molecules that caused these dye-blobs were efficiently removed by cleaning the PCR products over Performa DTR gel filtration cartridges (Fig. 1C) (16). These filtration cartridges removed dye molecules, nucleotides, and salts, after which the uptake of PCR products and a further increase in PH was observed (Fig. 1C).
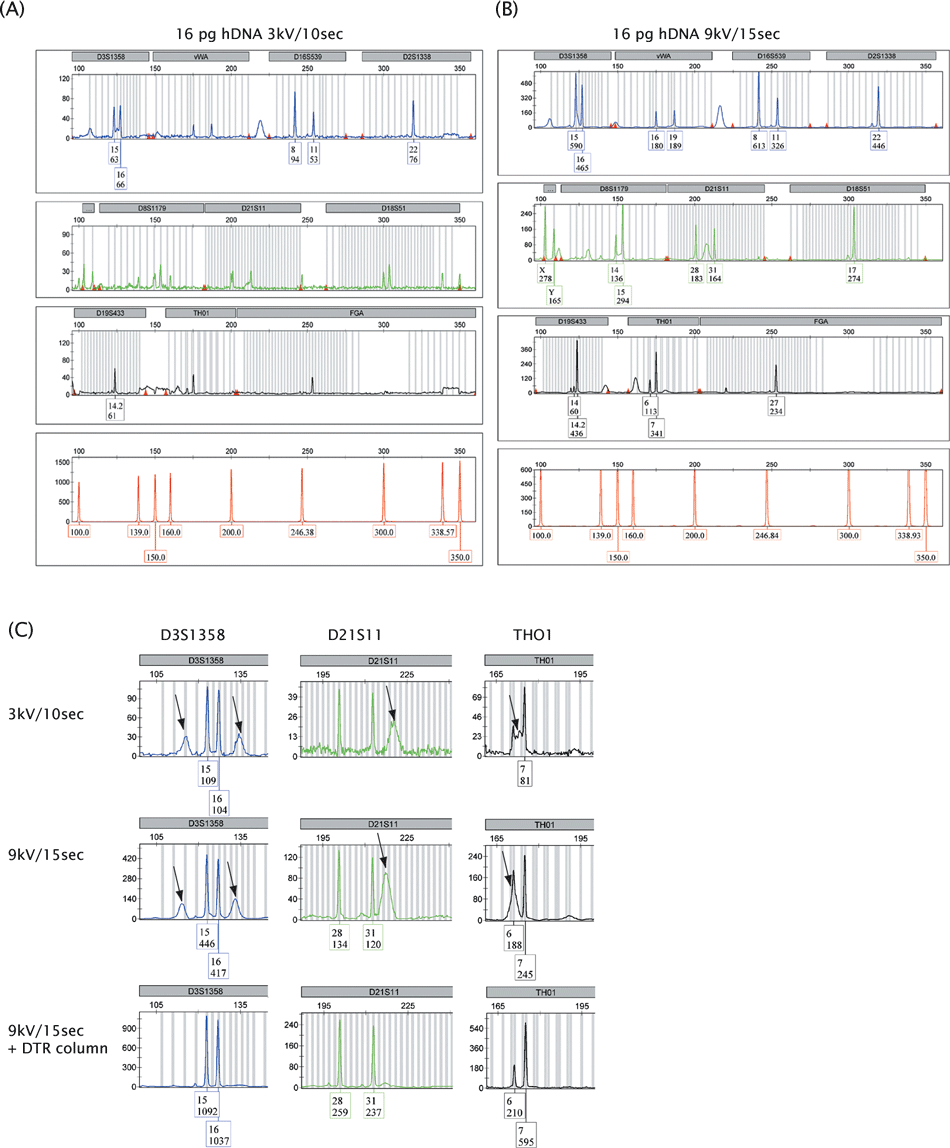
—Comparison of SGM+ profiles obtained using standard (A) and higher (B) injection settings. Polymerase chain reaction (PCR) was performed on 16 pg pristine DNA (hDNA). In the box underneath an allele, the upper number represents the allele call and the lower number, the peak height (in rfu). After boosted injection, all alleles except allele 19 in the FGA locus were detected. (C) Dye-blobs intensified upon boosted injection but could be removed using Performa DTR gel filtration cartridges. Dye-blobs in the loci D3S1358, D21S11, and THO1 are shown and indicated by arrows. The filtration cartridges removed dye molecules, nucleotides, and salts, which allowed the favored uptake of PCR products as was apparent from a further increase in peak heights. 164 mm × 177 mm (300 × 300 DPI).
For three pristine DNA samples (DNA 9947A, DNA 007, and hDNA), triplicate SGM+ PCRs with five different DNA inputs (125, 63, 31, 16, and 8 pg) were analyzed using standard 3 kV/10 sec injection and increased 9 kV/15 sec injection. While at normal injection settings, 125 pg DNA was required to obtain a full SGM+ profile, for one-third of the PCRs with 31 pg DNA input (five diploid cell equivalents), complete profiles were obtained. Signal strength (measured in rfu) increased 6.1-fold (average of 515 alleles). The genotyping results generated by boosted injection were based on amplifications from low-level DNA inputs and accompanied by allele dropouts (mainly for lower DNA inputs) and heterozygous peak imbalance while allele drop-ins and increased −1 stutters occurred sporadically (mainly for higher DNA inputs) (Table 1). Therefore, we analyzed STR profiles generated by boosted injection using standard marker-specific stutter ratios. As boosted injection aimed to obtain a maximum of genotyping information, we did not make use of an analysis threshold to prevent false homozygote calling. Figure 2 shows the average of the SGM+ genotyping results in the triplicate PCRs of the various DNA inputs using standard and increased injection. Incomplete but informative profiles (genotyping data for at least 50% of the SGM+ STR loci) were obtained for the amplifications using 8 or 16 pg DNA input (1, 2). Missing alleles occurred predominantly at the larger amplicons (results not shown).
| Method | No. Dropouts | No. Drop-ins | Heterozygote Balance |
|---|---|---|---|
| 3 kV, 10 sec | 0 (n = 45 profiles) | 0 (n = 45 profiles) | 0.69 ± 0.19 (n = 182 pairs) |
| 9 kV, 15 sec | 94 (n = 45 profiles) | 13 (n = 45 profiles)* | 0.52 ± 0.23 (n = 110 pairs)† |
| 0 (125 pg) | 6 (125 pg) | ||
| 3 (63 pg) | 4 (63 pg) | ||
| 13 (31 pg) | 3 (31 pg) | ||
| 29 (16 pg) | 0 (16 pg) | ||
| 49 (8 pg) | 0 (8 pg) |
- *8 at −1 stutter position; 3 at +1 stutter position.
- †Only alleles added after 9 kV/15 sec analyses.
- Five amounts of DNA input (125, 63, 31, 16, and 8 pg) of three pristine DNAs (hDNA, DNA007, and DNA9947A) were used in triplicate polymerase chain reactions.
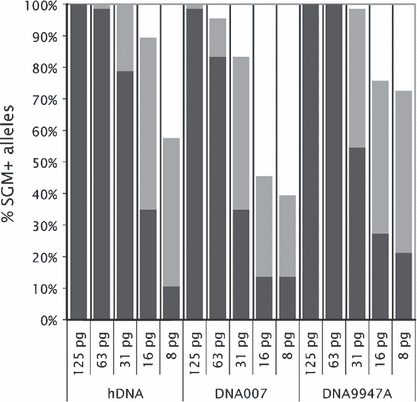
—Improved SGM+ genotyping data obtained by boosted capillary electrophoresis. Five DNA inputs (125, 63, 31, 16, and 8 pg) of three pristine DNAs (hDNA, DNA007, and DNA9947A) were used. Average percentages of detected alleles of triplicate polymerase chain reactions are presented. Dark gray bars indicate alleles called upon injection at 3 kV/10 sec; light gray bars correspond to additional alleles called after injection at 9 kV/15 sec. Missing alleles occurred predominantly at the larger amplicons (results not shown). 188 mm × 181 mm (300 × 300 DPI).
We studied the effects of boosted injection in SGM+ profiles in mock casework samples of known single donor origin that consisted of 228 positive samples and 108 negative controls (Table 2). Almost all of the positive samples with an incomplete profile upon standard injection showed an increase of concordant alleles when analyzed after boosted injection (on average 8.9 additional alleles per profile) (Table 2). Dropout alleles were found in 78.6% of the improved profiles (Table 2). Sporadic contamination was found in 2.6% of the standard DNA profiles and in 19.7% of the improved DNA profiles. Also in 22.2% of the negative samples, sporadic contamination was observed when using boosted injection, we inferred that low amounts of unrelated DNA were present on the items before usage by the volunteers (Table 2). Boosted injection effectively increased the sensitivity of STR typing in mock case-work samples. Improved profiles were obtained for various profiling methods like SGM+, Profiler, and Y-Filer and for samples of various origins (blood, saliva, sperm, skin epithelial, vaginal epithelial, and hair roots) that were collected by various sampling methods (cotton swap, nylon swap, tape lift, and laser micro dissection) (results not shown).
| No. Profiles | No. Among Positive Samples | No. Among Negative Samples |
|---|---|---|
| Samples | 228 | 108 |
| Incomplete 3 kV/10 sec | 204–6.0 detected alleles/profile | 0 |
| Improved 9 kV/15 sec | 201–8.9 additional alleles/profile | 0 |
| With sporadic contamination 3 kV/10 sec | 6 (2.6%)–1.7 alleles/profile | 0 |
| With sporadic contamination 9 kV/15 sec | 45 (19.7%)–1.7 alleles/profile* | 24 (22.2%)–2.3 alleles/profile |
| With dropout 3 kV/10 sec | 0 | 0 |
| With dropout 9 kV/15 sec | 158 (78.6%)–2.1 alleles/profile | 0 |
- *Average number of alleles determined among those profiles in which sporadic contamination was observed.
Higher Injection Settings Versus Increased PCR Cycle Number
The most frequently used low-level DNA profiling approach was increased PCR cycling by either performing 34 cycles on a fresh PCR or taking a portion of the 28 cycles PCR mixture, adding fresh polymerase, and amplifying for an additional six cycles (17). The 28 + 6 cycling approach had the advantage that both standard and high sensitivity genotyping data were obtained from a single DNA input. We compared triplicate PCRs of various DNA inputs (125, 63, 31, 16, and 8 pg) of three pristine DNA samples for standard 3 kV/10 sec injection, boosted 9 kV/15 sec injection, 34 PCR cycles, and 28 + 6 PCR cycles. Standard injection, boosted injection, and 28 + 6 cycling were performed on the same sample. Overloaded profiles with many pull-up peaks occurred for several DNA inputs especially when using elevated cycle number, and only the amplifications with DNA inputs of 16 and 8 pg could be analyzed using all three low-level DNA techniques. The average percentage of SGM+ alleles detected in the profiles generated by these three different techniques was quite similar (Fig. 3A). However, the average peak height (in rfu) was several folds lower with boosted injection than upon increased cycling (Fig. 3B). Hyperamplification was known to be accompanied by increased −1 stutter ratios, which was why elevated cycling genotyping data were analyzed using 1.5-fold locus-specific stutter ratios (boosted injection profiles are analyzed using the standard stutter ratios). All three low-level DNA methods were accompanied by a similar level of heterozygous peak imbalance (Table 3) and a similar frequency of allele dropouts (Table 3). However, allele drop-ins occurred more frequently in the profiles generated by increased cycling (Table 3). Most drop-in alleles represented +1 stutters (Table 3) which was most likely due to the higher amplification levels in the 34 and 28 + 6 cycling PCRs (14).
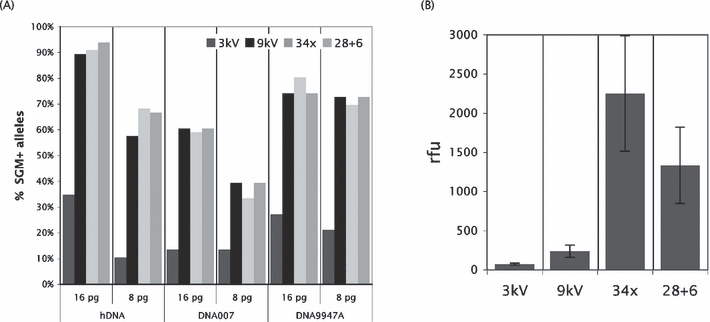
—(A) Boosted injection and increased polymerase chain reactions (PCR) cycle number resulted in a similar level of improvement of SGM+ genotyping. Average percentages of detected alleles of triplicate PCRs are presented. Dark gray bar, 3 kV/10 sec injection; black bar, 9 kV/15 sec injection; light gray bar, 34 amplification cycles; middle gray bar, 28 + 6 amplification cycles. Three pristine DNA samples (hDNA, DNA007, and DNA9947A) and two DNA inputs (16 and 8 pg) were used. (B) The average peak height (in rfu) was several fold higher when performing increased cycling. The data represented all alleles called in triplicate PCRs of both 16 and 8 pg DNA inputs. For each method, different numbers of allele calls were obtained: 3 kV/10 sec injection 61 alleles; 9 kV/15 sec injection 234 alleles; 34 amplification cycles 241 alleles, 28 + 6 amplification cycles 243 alleles. For boosted injection, peak heights increased 6.1-fold and for 28 + 6 cycling 35.0-fold (average of 61 alleles). 234 mm × 192 mm (300 × 300 DPI).
| Method | Heterozygote Balance | No. Dropouts | No. Drop-ins |
|---|---|---|---|
| 3 kV, 10 sec | 0.74 ± 0.08 (n = 6 pairs) | 0 | 0 |
| 9 kV, 15 sec | 0.58 ± 0.11 (n = 58 pairs)* | 77 (n = 18 profiles) | 0 |
| 34x | 0.54 ± 0.12 (n = 71 pairs) | 60 (n = 18 profiles) | 5 (n = 18 profiles)† |
| 28 + 6 | 0.55 ± 0.13 (n = 63 pairs)* | 72 (n = 18 profiles) | 5 (n = 18 profiles)‡ |
- *Only alleles added after 9 kV/15 sec or 28 + 6 cycling.
- †1 at −1 stutter position; 3 at +1 stutter position; 1 at other position.
- ‡1 at −1 stutter position; 4 at +1 stutter position.
- Two amounts of DNA input (16 and 8 pg) of three pristine DNAs (hDNA, DNA007, and DNA9947A) were used in triplicate polymerase chain reactions.
Higher Injection Settings to Enhance Detection of theMinor Component in Mixtures
We studied the effect of boosted injection versus more amplification rounds for the ability to improve genotyping of the minor component in mixtures. We used triplicate PCRs of 10:1 admixed samples of DNA9947A and hDNA with DNA inputs of 630 pg + 63 pg, 310 pg + 31 pg, 160 pg + 16 pg, and 80 pg + 8 pg. Eighteen of the 22 alleles in a SGM+ profile were distinct for hDNA in comparison to DNA9947A. Upon boosted injection, the major contributor induced overloaded profiles for the 630 pg + 63 pg and 310 pg + 31 pg DNA inputs. With more PCR cycles, all mixtures became over-amplified as was apparent from the occurrence of many pull-up peaks, but we analyzed the 80 pg + 8 pg mixtures notwithstanding the presence of these pull-up peaks (Fig. 4). Full profiles were obtained for the major contributor (DNA9947A, input 80 pg) for all three low-level DNA methods. Boosted injection revealed a similar number of detected alleles of the minor component as 34 and 28 + 6 cycling did (Table 4, Fig. 4). Boosted injection was found to result in less allele drop-ins (Table 4, Fig. 4). Three alleles of the minor component were at stutter position of an allele of the major component (Table 5), and may therefore be masked. This was only observed in profiles obtained after elevated cycling.
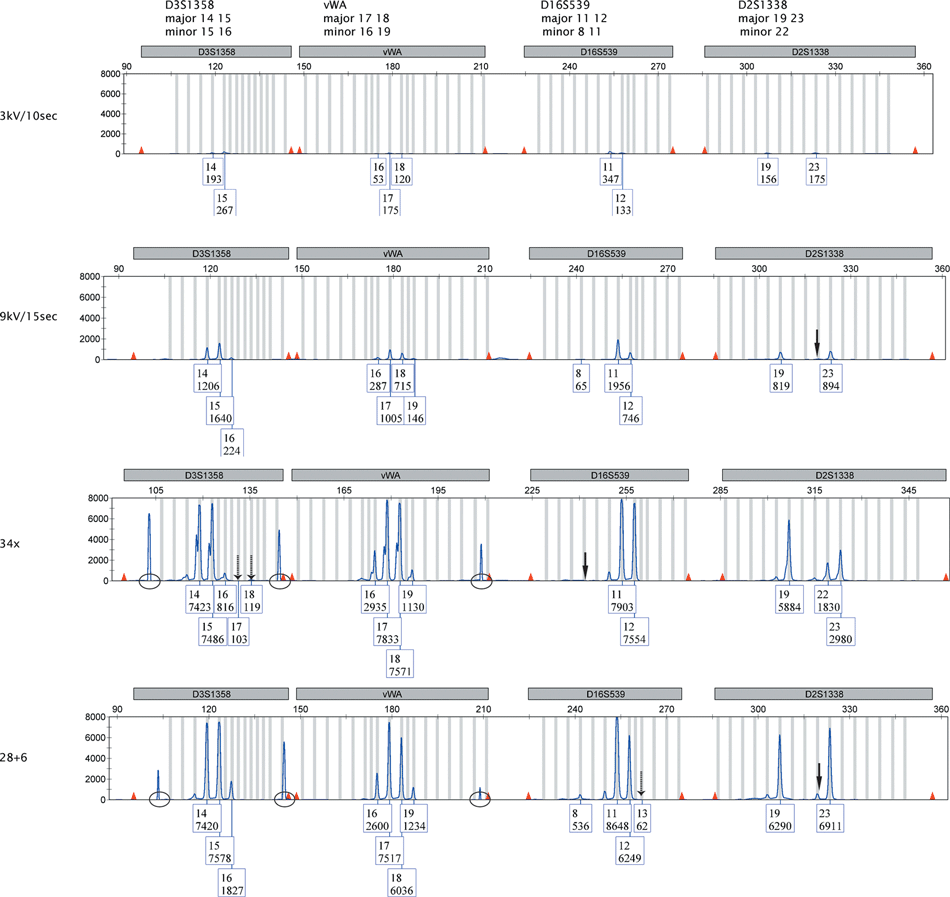
—Comparison of SGM+ genotyping results obtained for a 10:1 admixed sample consisting of 80 pg DNA9947A and 8 pg hDNA for standard DNA analysis and three low-level DNA techniques. Genotyping data are shown for the loci D3S1358, vWA, D16S539, and D2S1338 present in the FAM channel. Full arrows indicate the position of a dropout allele of the minor component. Dashed arrows indicate drop-in alleles. Circles indicate pull-up peaks. In the box underneath an allele, the upper number represents the allele call and the lower number the peak height (in rfu). 209 mm × 297 mm (300 × 300 DPI).
| Method | Input DNA9947A + hDNA | Alleles Major* | Alleles Minor† | Drop-Ins/ Per Profile |
|---|---|---|---|---|
| 3 kV | 160 pg + 16 pg | 22/22 | 8.3/18 | 0 |
| 3 kV | 80 pg + 8 pg | 21/22 | 2.7/18 | 0 |
| 9 kV | 160 pg + 16 pg | 22/22 | 14/18 | 1 |
| 9 kV | 80 pg + 8 pg | 22/22 | 11/18 | 0 |
| 34x | 80 pg + 8 pg | 22/22 | 14.7/18‡ | 4.7§ |
| 28 + 6 | 80 pg + 8 pg | 22/22 | 11/18‡ | 2.7¶ |
- Average of triplicate polymerase chain reaction is presented.
- *Calculated over all 22 SGM+ alleles.
- †Calculated over 18 alleles for which minor component differs from major component.
- ‡Occasionally alleles of minor contributor at stutter position removed manually upon using c. 15% stutter ratios.
- §2.0 at −1 stutter position; 1.3 at +1 stutter position; 1.3 at other positions.
- ¶0.3 at −1 stutter position; 1.3 at +1 stutter position; 0.3 at other positions.
| Sample | D3 | vWA | D16 | D2 | AMEL | D8 | D21 | D18 | D19 | THO1 | FGA |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Major | 14 15 | 17 18 | 11 12 | 19 23 | X | 13 | 30 | 15 19 | 14 15 | 8 9.3 | 23 24 |
| Minor | 15 16 | 16 19 | 8 11 | 22 | XY | 14 15 | 28 31 | 17 | 14 14.2 | 6 7 | 19 27 |
| Specific* | 16 | 16 19 | 8 | 22 | Y | 14 15 | 28 31 | 17 | 14.2 | 6 7 | 19 27 |
| 16 pg 3 kV, #1 | – | 16 19 | 8 | – | – | 15 | – | 17 | – | 6 | 19 |
| 16 pg 3 kV, #2 | – | – | – | 22 | Y | 15 | 28 | 17 | 14.2 | – | 19 27 |
| 16 pg 3 kV, #3 | – | 16 19 | – | 22 | – | – | 28 | 17 | – | – | – |
| 16 pg 9 kV, #1 | 16 | 16 19 15† | 8 | – | – | 14 15 | 28 31 26.2 | 17 | 14.2 12 | 6 7 | 19 |
| 16 pg 9 kV, #2 | – | 19 15 | 8 | 22 21 | Y | 14 15 | 28 31 | 17 | 14.2 | 6 | 19 27 |
| 16 pg 9 kV, #3 | – | 16 19 | – | 22 | Y | 14 | 28 | 17 | – | 6 7 | 19 27 |
| Consensus | – | 16 19 15 | 8 | 22 | Y | 14 15 | 28 31 | 17 | 14.2 | 6 7 | 19 27 |
| 8 pg 3 kV, #1 | – | – | – | 22 | – | – | – | 17 | – | – | 19 |
| 8 pg 3 kV, #2 | – | 16 | – | – | – | – | – | – | – | – | – |
| 8 pg 3 kV, #3 | – | – | – | – | – | – | – | 17 | – | – | – |
| 8 pg 9 kV, #1 | – | 19 | – | 22 | Y | 14 15 | 28 31 | 17 | – | 7 | 19 |
| 8 pg 9 kV, #2 | 16 | 16 19 | 8 | – | Y | 14 15 | 31 | – | – | 6 7 | 19 27 |
| 8 pg 9 kV, #3 | – | 19 | – | 22 | – | 14 15 | 29 | 17 | – | 7 | |
| Consensus | – | 16 19 | – | 22 | Y | 14 15 | 31 | 17 | – | 7 | 19 |
- AMEL, amelogenin.
- *Specific alleles for minor contributor.
- †Drop-in alleles are shown in italics.
- Polymerase chain reactions were performed in triplicate. Two DNA inputs (160 pg + 16 pg and 80 pg + 8 pg of DNA9947A + hDNA) were used.
We generated consensus profiles for the 160 pg + 16 pg and 80 pg + 8 pg mixtures after boosted injection (Table 5). The 160 pg + 16 pg consensus profile contained 17 of the 18 distinct alleles of the minor component plus one drop-in allele and the 80 pg + 8 pg consensus profile comprised 11 of the 18 distinct alleles (Table 4). We inferred that boosted injection could be of use to obtain more genotyping data on a minor component consisting of 8 or 16 pg DNA in 10:1 admixed samples, which was not achieved by using 28 + 6 or 34 amplification rounds as the samples get over-amplified. We expected more robust improvements from the use of higher injection settings when analyzing more evenly balanced mixtures (e.g., 5:1 or 2:1 mixture ratios, with a maximum DNA input of 100 pg).
Concluding Remarks
In this study we have shown that incomplete STR profiles can be efficiently improved by increasing the capillary electrophoresis injection settings on an ABI3130XL from 3 kV/10 sec to 9 kV/15 sec. Peak heights (in rfu) increased on average 6-fold. Amplification using six additional PCR cycles increased the peak height on average 35-fold. Interestingly, the percentage of alleles detected in samples with 16 or 8 pg DNA input was similar upon boosted injection or more PCR cycles, although the average peak height was much lower in the 9 kV/15 sec injection profiles than in those generated with an elevated cycle number. As dye-blobs were also enhanced upon boosted injection, effort might be needed to distinguish true peaks from dye-blobs or remove dye molecules from PCR mixtures, which was hardly an issue in 34-cycle or 28 + 6 cycle profiles. On the other hand, the use of 34 or 28 + 6 amplification rounds could result in overloaded profiles with many pull-up peaks, a feature hardly observed upon boosted injection. As with other low-level DNA methods, artifacts occurred due to stochastic effects during amplification. These included heterozygous peak imbalance, allele dropouts, and allele drop-ins. Occurrence of increased stutter peaks was rare which implied that boosted injection (unlike elevated cycling) did not necessitate the use of increased stutter ratios. Amplification of pristine DNAs showed that boosted injection and more PCR cycles resulted in a similar level of heterozygous peak imbalance and number of dropouts in the genotyping data. The main difference between the two approaches was the presence of more allele drop-ins (like +1 stutters) when using additional amplification rounds. The occurrence of allele drop-ins in boosted injection profiles seemed to depend on the type of sample because for pristine DNAs no drop-ins were found while in the majority of our mock casework samples a few alleles due to sporadic contamination were detected. We inferred that the sampled items were not completely free of sporadic DNA contamination before usage by our volunteers.
We were able to improve the genotyping data of the minor component in 10:1 admixed samples by boosted injection, while amplification of these mixtures for 34 or 28 + 6 cycles resulted in over-amplified samples and overloaded profiles that would normally not be analyzed. We did not observe masking of alleles of the minor component in the 10:1 admixed samples that were at stutter position of an allele of the major component.
Clearly, boosted injection had to be considered a low-level DNA technique that was accompanied by a substantial number of allele dropouts. We recommend generating a consensus profile from multiple independent amplifications. In our laboratory, a consensus profile consisted of all alleles detected with the standard procedure plus those alleles that were detected in at least two out of three profiles generated by a low-level DNA procedure. The low-level DNA technique of 9 kV/15 sec capillary electrophoresis injection had the advantage that for each separate PCR, both a standard and an enhanced STR profile were easily obtained. In case where higher peaks were required, one could decide to perform 28 + 6 cycling subsequently (17). This could be carried out on the same PCR product without using more DNA extract.
In summary, boosted capillary electrophoresis is a simple method to increase the sensitivity of STR typing. It is accompanied by the occurrence of allele dropouts and heterozygous peak imbalance, but does not require the use of increased stutter ratios. Boosted injection is suited to improve not only single donor profiles but also the genotyping data of the minor component in mixtures. The method has been accredited for case work in our laboratory.
Acknowledgments
We thank Fleur Beemster and Loes Schoenmakers for technical assistance. We thank Ate Kloosterman and Simone van Soest for critically reading the manuscript.




