The Phylogeny of Colpodellids (Alveolata) Using Small Subunit rRNA Gene Sequences Suggests They are the Free-living Sister Group to Apicomplexans
The GenBank accession numbers for new SSU rDNA sequences: Colpodella pontica (AY078092) and Colpodella sp., ATCC 50594 (AY142075).
Abstract
ABSTRACT. In an attempt to reconstruct early alveolate evolution, we have examined the phylogenetic position of colpodellids by analyzing small subunit rDNA sequences from Colpodella pontica Myl'nikov 2000 and Colpodella sp. (American Type Culture Collection 50594). All phylogenetic analyses grouped the colpodellid sequences together with strong support and placed them strongly within the Alveolata. Most analyses showed colpodellids as the sister group to an apicomplexan clade, albeit with weak support. Sequences from two perkinsids, Perkinsus and Parvilucifera, clustered together and consistently branched as the sister group to dinoflagellates as shown previously. These data demonstrate that colpodellids and perkinsids are plesiomorphically similar in morphology and help provide a phylogenetic framework for inferring the combination of character states present in the last common ancestor of dinoflagellates and apicomplexans. We can infer that this ancestor was probably a myzocytotic predator with two heterodynamic flagella, micropores, trichocysts, rhoptries, micronemes, a polar ring, and a coiled open-sided conoid. This ancestor also very likely contained a plastid, but it is presently not certain whether it was photosynthetic, and it is not clear whether extant perkinsids or colpodellids have retained the organelle.
INTRACELLULAR parasitism, phototrophy, and predation are among the most dissimilar modes of eukaryotic life, yet each of these specializations is represented in three rather closely related groups of unicellular organisms: apicomplexans, dinoflagellates, and ciliates. Morphological and molecular characters indicate that these extremely divergent groups of eukaryotes share a common ancestor to the exclusion of all other major groups, collectively they are called the Alveolata. Synapomorphies for alveolates include a system of inner membranes called “alveoli” and distinct openings in the cell surface called “micropores” (Patterson 1999; Siddall et al. 1997); members of the group also possess features present in many other eukaryotes such as tubular mitochondrial cristae and, in general, a closed mitosis. Despite these ultrastructural similarities, the differences in morphology and life history between apicomplexans, dinoflagellates, and ciliates are profound. Consequently, inferences about the ancestral states and intermediate stages that led to these extremely varied lineages are based more on speculation than rigorous empirical analyses.
It is becoming more apparent, however, that there are a variety of alveolates that do not fit neatly within the three major groups discussed above, such as Acrocoelus, Colpodella, Colponema, Cryptophagus, Oxyrrhis, Parvilucifera, and Perkinsus (Brugerolle 2002; Dodge and Crawford 1971; Fernandez et al. 1999; Mignot and Brugerolle 1975; Myl'nikov 1991, 2000; Noren et al. 1999; Perkins 1976, 1996; Saldarriaga et al. 2002; Siddall et al. 1997, 2001; Simpson and Patterson 1996). The combination of features in these taxa suggest that they may be very important for inferring specific states in ancestral alveolates and for understanding the early evolution of apicomplexans, dinoflagellates, and ciliates. For instance, evidence suggests that perkinsids (Cryptophagus, Parvilucifera, and Perkinsus), which are parasites of mollusks and microeukaryotes, are the earliest diverging sister lineages to dinoflagellates (Ellis et al. 1998; Goggin and Barker 1993; Noren et al. 1999; Reece et al. 1997; Saldarriaga et al. 2002; Siddall et al. 1997). The zoospores of perkinsids have two dissimilar flagella and possess structures traditionally attributed only to apicomplexans such as a microtubular conoid-like apparatus, rhoptries, and micronemes (Azevedo 1989; Azevedo et al. 1990; Coss et al. 2001; Levine 1978; Perkins 1976, 1996). These comparative data provide valuable clues for inferring the characteristics of the last common ancestor of apicomplexans and dinoflagellates.
Colpodellids are biflagellated predators with a four-way divisional cyst and have a suite of features that are remarkably similar to perkinsids. (Myl'nikov 1991, 2000; Simpson and Patterson 1996) Understanding the phylogeny of colpodellids could shed considerable light on early alveolate evolution, but our understanding of their position within alveolates is hampered by conflicting data. Colpodella sp. (American Type Culture Collection 50594) has been shown to branch as an early lineage of apicomplexans when a partially unresolved SSU rDNA sequence was analyzed with parsimony and to be the sister lineage to ciliates when this SSU rRNA was concatenated with the DNA sequence of an actin gene (Siddall et al. 2001). The latter result led the authors to conclude that the ancestral alveolate possessed the character states shared by Colpodella and Perkinsus (Siddall et al. 2001). However, it has been subsequently shown that the actin sequence used in this analysis was misattributed to Colpodella sp. (Saldarriaga et al. 2002) and was likely amplified from the euglenozoan prey organism, Bodo. Altogether, the phylogenetic position of colpodellids within alveolates and the associated interpretations about alveolate ancestry remain unclear.
The hypothetical framework we are using to comprehend early alveolate evolution is outlined in Fig. 1. We recognize two separate questions: (1) What were the biological properties of the most recent common ancestor of apicomplexans and dinoflagellates (A in 1, 2) what were the properties of the most recent common ancestor of all alveolates (B in Fig. 1)? If the earliest diverging sister lineages of apicomplexans (C in Fig. 1) and dinoflagellates (D in Fig. 1) are very similar on a character state-by-state basis, then an extraordinarily confident inference could be made about the biological features of their common ancestor (A in Fig. 1). Likewise, if there are many character states shared between the earliest diverging sister lineage to ciliates (E in Fig. 1) and the inferred ancestor of apicomplexans and dinoflagellates (A in Fig. 1), then an uncommonly robust inference can be made about the biological features of the most recent common ancestor of all alveolates (B in Fig. 1). In order to make these potentially powerful inferences, we must identify the “key” taxa and use appropriate data and methods to properly place them into a robust phylogenetic context.
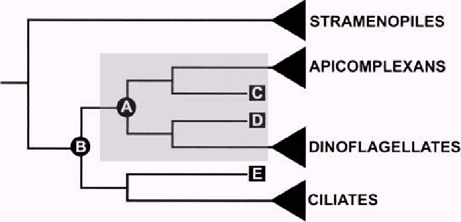
Hypothetical framework for inferring character states in ancestral alveolates. The three main groups of alveolates (apicomplexans, dinoflagellates, and ciliates) have very different modes of life and are extraordinarily divergent morphologically (as indicated by triangles), yet they are united by the presence of a distinct system of inner membranes (alveoli), micropores, and molecular sequence characters (Fast et al. 2002). Many extant alveolates that do not fit neatly within these three groups (lineages C, D, and E) may shed considerable light on two key questions: (1) What were the character states present in the most recent common ancestor of dinoflagellates and apicomplexans (extinct ancestor A) and (2) what were the character states present in the most recent common ancestor of all alveolates (extinct ancestor B)? If the earliest diverging extant sister lineage to apicomplexans (C) is strikingly similar on a state-by-state basis to the earliest diverging extant sister lineage to dinoflagellates (D), then unprecedented inferences can be made about the character states of ancestor A as indicated by the shaded box. Likewise, a comparative analysis of the earliest diverging extant sister lineage to ciliates (lineage E) with lineages C and D should provide important insights into the character states present in the extinct common ancestor of all alveolates (ancestor B). A better understanding of the biology and phylogenetic positions of these key taxa (lineages C, D, and E) will eventually lead to a more confident and accurate explanation for the origins of plastids and intracellular parasitism within alveolates.
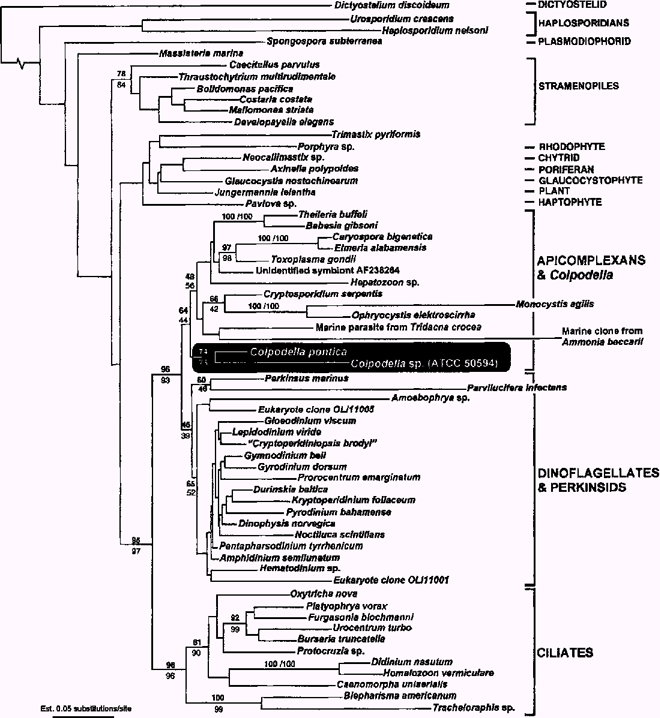
Gamma-corrected ML tree (–InL = 22447) derived from a matrix of 62 SSU rDNA sequences and 1431 sites showing the phylogenetic position of Colpodella sp. and C. pontica (shaded) among major eukaryotic groups (bracketed to the right). Numbers above relevant branches correspond to gamma-corrected bootstrap percentages from weighted neighbour-joining (bold) and Fitch-Margoliash (plain). The two Colpodella sequences clustered together in all analyses with moderate support (74/73) and were strongly positioned within the alveolates (95/97), where apicomplexans formed their sister group. This position was not affected by the inclusion of extraordinary divergent sequences from Oxyrrhis, Plasmodium, and Gregarina in ML and MP analyses. Transition/transversion ratio was 1.95.
In this vein, we have attempted to demonstrate the phylogenetic position of colpodellids using a SSU rDNA sequence from the predatory alveolate Colpodella pontica (Mylni'kov 2000) and a new SSU rDNA sequence from Colpodella sp. (American Type Culture Collection 50594). Comparative sequence analyses of these data provide evidence for the monophyly of colpodellids and their sisterhood to the “true” apicomplexans. This position of colpodellids within alveolates combined with the relatively well-established sister relationship between perkinsids and dinoflagellates provides an ideal phylogenetic framework for inferring the character states present in the common ancestor of apicomplexans and dinoflagellates.
MATERIALS AND METHODS
Collection and culture conditions. Colpodella pontica Myl'nikov 2000 was isolated from the coastal waters of the Black Sea near Yalta (Crimea, Ukraine) in December 1987. Cultures of C. pontica were maintained in a Schmaltz-Pratt medium (add the following to 1 L of water: 28.15 g NaCl, 0.67 g KCl, 5.51g MgCl2·6H2O, 6.92 g MgSO4·7H2O, 1.45 g CaCl2·H2O, 0.1 g KNO3, 0.01 g K2HPO4·3H2O) with a salinity adjusted to 20–22‰ and inoculated with Escherichia coli and a eukaryotic prey organism, Bodo sorokini Zhukov 1975 Myl'nikov 2000). Colpodella pontica can be obtained from the culture collection maintained at the Institute for Biology of Inland Waters, Russian Academy of Sciences.
Colpodella sp. was obtained from the American Type Culture Collection (ATCC 50594) (Manassas, Virginia, USA) and maintained in ATCC culture medium 802 (Sonneborn's Paramecium medium) inoculated with Klebsiella pneumoniae and a eukaryotic prey organism, Bodo caudatus.
DNA extraction, PCR amplification, and sequencing. Genomic DNA was extracted from a pellet of C. pontica (including Bodo sorokini-prey) and from a pellet of Colpodella sp. including Bodo caudatus-prey) using a standard hexadecyltrimethylammonium bromide (CTAB) extraction protocol (Zolan and Pukkila 1986). Small subunit (SSU) rRNA genes were amplified as single fragments using universal eukaryotic primers and a standard PCR protocol (Leander et al. 2002). The PCR fragments from C. pontica and B. sorokini were different sizes (∼ 1,700 bp for C. pontica and > 2,000 bp for Bodo sp.) and clearly distinguishable on an agarose gel. Fragments from both size products were gel-isolated with an UltraClean 15 DNA purification Kit (MoBio Laboratories, Inc., Solana Beach, California, USA) and inserted into a pGEM-T vector (Promega Corporation, Madison, Wisconsin). The PCR fragments from Colpodella sp. (ATCC 50594) were gel-isolated and inserted into the pCR 2.1 vector using the TOPO TA cloning kit (Invitrogen, Frederick, Maryland, USA). One clone from each species of Colpodella was completely sequenced with ABI big-dye reaction mix using vector primers and four internal primers oriented in both directions.
Alignments and phylogenetic analysis. Three different alignments were constructed using MacClade 4 (Maddison and Maddison 2000). The new sequences from C. pontica and Colpodella sp. (ATCC 50594) were added to an existing alignment consisting of diverse alveolates and representative sequences from other major eukaryotic groups—(Leander et al. 2002) a 65-taxon alignment. However, we focused on two smaller alignments. In order to minimize artifacts of long branch attraction (LBA), three of the most divergent SSU rDNA sequences from the 65-taxon alignment (Oxyrrhis marina, Gregarina polymorpha, and Plasmodium falciparum) were excluded to form a 62-taxon alignment with 1,431 unambiguously aligned sites. A more comprehensive analysis of the position of Colpodella sp. within alveolates was achieved by trimming the number of taxa in the 62-taxon alignment to 32 ingroup taxa representative alveolates) and 3 outgroup taxa (representative stramenopiles)—a 35 taxon alignment containing 1,442 sites. These SSU rDNA sequence alignments are available upon request.
Maximum likelihood (ML) and distance methods under different DNA substitution models were performed on the alignments. The alpha shape parameters for site-to-site rate variation were estimated from the data using HKY and a gamma distribution with invariable sites and eight rate categories (0.29 for the 62-taxon alignment and 0.31 for the 35-taxon alignment; fraction of invariable sites was 0.00 for both alignments). For both the 62-taxon and 35-taxon datasets, corrected ML trees were constructed with PAUP* 4.0 using the general time reversible (GTR) model for base substitutions (Posada and Crandall 1998; Swofford 1999). For the 35-taxon dataset, ML bootstrap analyses were performed in PAUP (Swofford 1999) on one hundred resampled datasets under an HKY model using corrections for site-to-site rate variation estimated from the original dataset.
Distances for both SSU rDNA datasets were calculated with TREE-PUZZLE 5.0 using the HKY substitution matrix (Strimmer and von Haeseler, 1996) and with PAUP using the GTR model. Distance trees were constructed with minimum evolution (ME—using GTR) in PAUP, (Swofford 1999) weighted neighbor joining (WNJ) using Weighbor, (Bruno et al. 2000) and Fitch-Margoliash using FITCH (Felsenstein 1993) with global rearrangements and ten input order jumbles. One hundred bootstrap datasets were generated with SEQBOOT (Felsenstein 1993). Respective distances were calculated with the shell script “puzzleboot” (M. Holder and A. Roger, http://www.tree-puzzle.de) using the alpha shape parameter and transition/transversion ratios estimated from the original datasets. The number of input order jumbles was reduced to three in the Fitch-Margoliash analyses of the bootstrap distance trees.
Although misleading results due to LBA were expected, we also analyzed the 35-taxon dataset with parsimony. Nucleotides were treated as independent, unordered, character states of equal weight and gaps were treated as missing data. A heuristic search was performed using PAUP* 4.0 with ACCTRAN character state optimization, tree bisection reconnection (TBR) branch swapping, random step-wise addition of taxa, and MULTREES on. Non-parametric bootstrap values from 500 replicates were generated to evaluate the robustness of each node on the most parsimonious tree(s). The tree length, number of most parsimonious trees, number of informative characters, CI, and RI were reported.
GenBank accession numbers. (AF069516) Amoebophrya sp., (AF274256) Amphidinium semilunatum, (U43190) Axinella polypoides, (AF158702) Babesia gibsoni, (M97909) Blepharisma americanum, (AF167154) Bolidomonas pacifica, (U82204) Bursaria truncatella, (AF174368) Caecitellus parvulus, (U97108) Caenomorpha uniserialis, (AF060975) Caryospora bigenetica, (AY142075) Colpodella sp. (American Type Culture Collection 50594), (AY078092) Colpodella pontica, (X53229) Costaria costata, (AF080097) Cryptoperidiniopsis brodyi, (AF093502) Cryptosporidium serpentis, (U37107) Developayella elegans, (K02641) Dictyostelium discoideum, (U57771) Didinium nasutum, (AF239261) Dinophysis norvegica, (AF231803) Durinskia baltica (formerly Peridinium balticum), (AF291427) Eimeria alabamensis, (AJ402327) Eukaryote clone OLI11001, (AJ402349) Eukaryote clone OLI11005, (X65150) Furgasonia blochmanni, (X70803) Glaucocystis nostochinearum, (L13716) Gloeodinium viscum, (U37406) Gymnodinium beii, (AF274261) Gyrodinium dorsum, (X74131) Haplosporidium nelsoni, (AF286023) Hematodinium sp., (AF297085) Hepatozoon sp., (L26447) Homalozoon vermiculare, (X91784) Jungermannia leiantha, (AF274268) Kryptoperidinium foliaceum, (AF022199) Lepidodinium viride, (M87333) Mallomonas striata, (U07937) Marine clone misattributed to Ammonia beccarii, (AB000912) Marine parasite from Tridacna crocea, (AF174370) Massisteria marina, (AF457127) Monocystis agilis, (M59761) Neocallimastix sp., (AF022200) Noctiluca scintillans, (AF129883) Ophryocystis elektroscirrha, (M14601) Oxytricha nova, (AF133909) Parvilucifera infectans, (AJ243369) Pavlova sp., (AF022201) Pentapharsodinium tyrrhenicum, (AF126013) Perkinsus marinus, (AF060454) Platyophrya vorax, (AF136425) Porphyra sp., (Y16239) Prorocentrum emarginatum, (AF194409) Protocruzia sp., (AF274275) Pyrodinium bahamense, (AF310901) Spongospora subterranea, (AF236097) Theileria buffeli, (AB022111) Thraustochytrium multirudimentale, (M97703) Toxoplasma gondii, (L31520) Tracheloraphis sp., (AF244903) Trimastix pyriformis, (AF238264) Unidentified symbiont, (AF255357) Urocentrum turbo, (U47852) Urosporidium crescens
RESULTS AND DISCUSSION
Phylogeny of colpodellids. The 62-taxon alignment included representative SSU rDNA sequences from a variety of eukaryotic groups, and in all analyses, the two Colpodella sequences grouped together and clustered strongly within a clade of alveolates (BP 95/97, Fig. 2). More significantly, the Colpodella clade branched as the sister lineage to an apicomplexan clade in corrected ML (GTR model of substitution) and weighted-distance analyses, albeit with weak bootstrap support (2, 3). Moreover, when the extremely divergent sequences from Oxyrrhis marina, Plasmodium falciparum, and Gregarina polymorpha were included in corrected ML analyses (65-taxon dataset), the position of Colpodella did not change, which is evidence that the sister relationship of Colpodella with apicomplexans is stable despite potential artifacts from LBA. Distance analyses using minimum evolution and a GTR model of substitution produced the same basic topology as in Fig. 3 except that the Colpodella clade branched as the earliest diverging lineage in a clade consisting of the apicomplexans, dinoflagellates, and perkinsids. Parsimony analysis (CI = 0.44, RI = 0.43, number of most parsimonious trees = 24, tree length = 2623, number of informative characters = 447) appeared to be affected by LBA as the two longest branches in the analysis, Parvilucifera and the marine clone from Ammonia beccarii (GenBank U07937—previously shown to be a gregarine sequence, Leander et al. 2002), grouped together and showed a weak affinity to the Colpodella clade. Nonetheless, in corrected ML and distance analyses, Parvilucifera and Perkinsus formed a moderately supported group that diverged as the sister to a moderately supported dinoflagellate clade (2, 3), which is consistent with most previous studies (Ellis et al. 1998; Goggin and Barker 1993; Leander et al. 2002; Reece et al. 1997; Saldarriaga et al. 2002; van de Peer and de Wachter 1997).
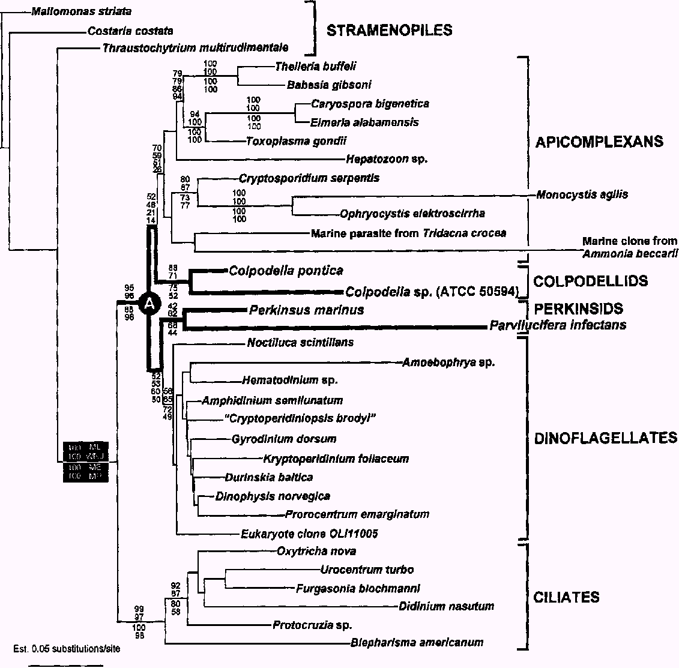
Gamma-corrected ML tree (– InL = 14324) inferred from an alignment of 35 SSU rDNA sequences and 1442 sites, with stramenopiles as the outgroup, showing the phylogenetic position of Colpodella, Perkinsus, and Parvilucifera within alveolates. The numbers at the branches denote gamma-corrected bootstrap percentages using each of the following methods (from top to bottom): maximum likelihood—using GTR (ML), weighted neighbor-joining (WNJ), and minimum evolution—using GTR (ME). The lowermost numbers refer to bootstrap percentages using parsimony (MP). Except in parsimony analyses, colpodellids and perkinsids were consistently positioned as the earliest diverging sister lineages to apicomplexans and dinoflagellates, respectively, with low support. The shared character states present in colpodellids and perkinsids are inferred to have been retained from the states present in their most recent common ancestor (A), which is indicated by heavy lines (See text for discussion). Transition/transversion ratio was 1.95.
Reconstructing the common ancestor of apicomplexans and dinoflagellates. A robust inference about the biological features of the most recent common ancestor of all alveolates remains elusive. However, significant progress is being made in identifying extant lineages that help us reconstruct the ancestor of apicomplexans and dinoflagellates using molecular sequence comparisons and comparative morphological approaches (Brugerolle 2002; Goggin and Barker 1993; Myl'nikov 1991, 2000; Noren et al. 1999; Perkins 1996; Reece et al. 1997; Saldarriaga et al. 2002; Siddall et al. 1997, 2001; Simpson and Patterson 1996). Although protein sequences are still necessary to maximize confidence in the phylogenetic position of Colpodella, analysis of SSU rDNA provides tantalizing evidence that colpodellids are the earliest diverging sister group to apicomplexans. On the other hand, SSU and several protein phylogenies all consistently show that perkinsids are the earliest diverging sister group to dinoflagellates (Saldarriaga et al. 2002). Together, this allows us to rebuild the most recent common ancestor of dinoflagellates and apicomplexans more confidently by looking closely at the range of character states shared by perkinsids and colpodellids.
It is possible to infer that die ancestor of apicomplexans and dinoflagellates had tubular mitochondrial cristae, trichocysts, micropores, and two heterodynamic flagella inserted either laterally as in Colpodella pugnax Cienkowski 1865, Perkinsus atlanticusAzevedo 1989, and Oxyrrhis marina (Azevedo 1989; Dodge and Crawford 1971; Simpson and Patterson 1996) or subapically as in Cryptophagus (Brugerolle 2002) and Colpodella perforans (formerly named Spiromonas perforans) (Brugerolle and Mignot 1979). The anterior flagellum was probably decorated with simple mastigonemes, but the presence of mastigonemes in colpodellids and robust inferences about the characteristics of the common ancestor of all alveolates still need to be established. The last ancestor of dinoflagellates and apicomplexans almost certainly possessed an apparatus resembling the apical complex of “true” apicomplexans represented by an open-sided conoid, rhoptries, micronemes and, possibly, the apical polar ring. It is very likely that this apical apparatus enabled the ancestral predators to access the cytoplasm of other cells.
An open-sided conoid that is associated with rhoptries and micronemes appears to be a synapomorphy for the dinoflagellate-apicomplexan clade and therefore, from a phylogenetic perspective, may be regarded as a “defining feature” of the group. The conoid is problematic as a diagnostic feature because it has been modified into a closed conoid in apicomplexans and apparently lost in dinoflagellates. However, either the microtubular baskets in the peduncle of some phagotrophic dinoflagellates such as Amphidinium poecilochroum (Larsen, 1988) or the “apical pore complex” might be derived from open-sided conoids. The “pseudoconoids” (small c-shaped microtubular sheets) of Parvilucifera and Cryptophagus may be an intermediate state in the transformation from a more robust open-sided conoid to the microtubular baskets of dinoflagellates. Nonetheless, the dinoflagellate-apicomplexan clade is not synonymous with the paraphyletic “Apicomonada”sensuCavalier-Smith (1993) or the “Perkinsozoa” sensu Noren (1999) as it includes not only perkinsids and colpodellids but also all apicomplexans and dinoflagellates; the former name could more informatively tag this more inclusive grouping in place of the name “Miozoa” (Cavalier-Smith 1993, 1999).
There are a variety of other features of the last ancestor of dinoflagellates and apicomplexans that are more difficult to interpret. For instance, the morphology and organization of alveoli are not yet inferable because we do not know enough about these features in colpodellids and perkinsids. It is possible that the common ancestor of all alveolates and the last ancestor of dinoflagellates and apicomplexans possessed a ventral groove that is homologous to the feeding groove emphasized by the “excavate hypothesis” (Simpson and Patterson 1999). Quite a few enigmatic alveolates possess distinct ventral grooves, such as Colponema, Acrocoelus, and many species of Colpodella (Fernandez et al. 1999; Mignot and Brugerolle, 1975; Myl'nikov 1991; Simpson and Patterson 1996), and it may even turn out that the sulcus of dinoflagellates is homologous to these grooves. However, confidence in these hypotheses awaits more research on a number of key taxa. Similarly, we do not know whether the four-way divisional cyst (the defining feature of colpodellids) was present in the last ancestor of apicomplexans and dinoflagellates, but molecular sequences from enigmatic alveolates such as Cryptophagus subtilisBrugerolle 2002 and Colpodella perforansSimpson and Patterson 1996 may provide some necessary data. Both are parasites of the cryptomonad “Chilomonas” and are strikingly similar in overall cell morphology, but Colpodella perforans (as the genus name implies) possesses a four-way divisional cyst and Cryptophagus subtilis does not (Brugerolle 2002; Brugerolle and Mignot 1979). If these taxa turn out to be more closely related to perkinsids (and dinoflagellates) than to colpodellids, then colpodellids as a group defined by the four-way divisional cyst would become paraphyletic and encompass the last ancestor of dinoflagellates and apicomplexans.
Lastly, it is well-known that apicomplexans contain a vestigial plastid (Kohler et al. 1997; McFadden 2000; McFadden et al. 1996), and molecular evidence indicates that this “apicoplast” is not only homologous to the photosynthetic, peridinin-containing plastids of many dinoflagellates (Fast et al. 2001; Zhang et al. 2000) but also the plastids of stramenopiles (Fast et al. 2001). These data suggest that ancestors A and B contained plastids of some kind, and it is interesting to speculate as to whether or not these plastids were photosynthetic. Ancestor A may have been an active phototroph, which would mean that photosynthesis was lost independently in perkinsids and the lineage leading to colpodellids and apicomplexans. While this may demand several parallel losses, it is clear from examining dinoflagellates and stramenopiles that plastid loss is a relatively common event in these lineages (Saldarriaga et al. 2001). An alternative possibility is that an active, photosynthetic plastid like those found in stramenopiles was reduced in the ancestor of all alveolates (ancestor B) and that photosynthesis was regained in dinoflagellates. While this may seem unlikely at first glance, it is important to note that dinoflagellates are remarkably promiscuous in acquiring plastids: the peridinin-containing plastid has been independently replaced in various lineages by plastids derived from haptophytes, cryptophytes, stramenopiles, and green algae (Saldarriaga et al. 2001; Delwiche 1999; Tengs et al. 1999). Therefore, it is not far-fetched to suspect that the peridinin-containing plastid of dinoflagellates may have arisen from a tertiary endosymbiosis with a plastid from either a haptophyte, cryptophyte, or stramenopile. Distinguishing between these alternatives will require a better understanding of the evolutionary relationship between the peridinin-containing dinoflagellate plastid and the apicomplexan plastid.
ACKNOWLEDGMENTS
We wish to thank J. F. Saldarriage and J. M. Archibald for comments on the manuscript. O.N.K. was supported by the Russian Foundation for Basic Research (02–04–48958, 02–04–48265, 00–15–97905). B.S.L. was supported by the National Science Foundation (Postdoctoral Research Fellowship in Microbial Biology, USA). This work was supported by a grant to P.J.K. from the Canadian Institutes for Health Research (MOP-42517). P.J.K. is a Scholar of the Canadian Institute for Advanced Research and the Michael Smith Foundation for Health Research.




