Mechanisms of Amino Acid and Glucose Uptake by Pneumocystis carinii
Amino acids serve as important precursors for the synthesis of proteins as well as other compounds such as lipids. Although it is not known whether Pneumocystis carinii requires exogenous glucose as a carbon source for energy production, there is evidence that this sugar is needed for axenic culture growth (A. Clarkson, S. Merali, pers. common.). While the uptake and incorporation of radiolabeled and fluorescent lipids by P. carinii have been studied [5], transport kinetics for only palmitate and oleate have been reported [7]. Transport of these fatty acids exhibited first order kinetics, suggesting that the organism has at least one fatty acid transporter system. With the exception of S-adenosylmethionine (SAM) transport [6], the mechanisms by which the organism takes up different water-soluble nutrients have not been examined. Thus, we analyzed the kinetic parameters and the effects of a number of different types of inhibitors to determine what kinds of mechanisms were involved in amino acid and glucose uptake by the organism.
MATERIALS AND METHODS
Preparations (>95%–100% purity; 80%–95% viability) of P. carinii organisms were isolated from infected rat lungs as previously described [5]. After the 2–3 h isolation and purification protocol, organisms were immediately incubated with radiolabeled compounds. The organisms were suspended in a buffered solution containing 100 mM HEPES buffer (pH 7.4), 1.8 mM CaCl2, and 145 mM NaCl. For amino acid uptake assays, the incubation medium was supplemented with 2% glucose. Each aliquot of organisms contained approximately 4 × 107 cells and uptake was measured at 37°C for various periods and substrate concentrations. Cells were washed twice and the radioactivity in the cell pellet was quantified. Uptake was expressed as pmol/mg total organism protein/time. Uptake kinetics was also analyzed in experiments that included one or more of a variety of inhibitor compounds.
Radiolabeled glucose uptake was examined and compared with that of the nonmetabolizable analog 2-deoxyglucose (2-DOG). Detailed analyses were performed using 2-DOG.
RESULTS AND DISCUSSION
Amino acid uptake
The time course of aspartic acid uptake by P. carinii was linear up to 45 min using an incubation medium containing 1 mM of the substrate, thus assays using different substrate concentrations were performed using 30-min incubations. Uptake of aspartic acid obeyed Picks Law when tested in media containing up to 5 mM aspartate at pH 7.4 and up to 20 mM aspartate at pH 5.5 (Fig. 1). The direct relationship between uptake rate and substrate concentration with no evidence of saturation demonstrated that simple diffusion was the mechanism by which P. carinii translocated this acidic amino acid. The Q10 of aspartate uptake was 1.10, which is consistent with strictly physical processes.
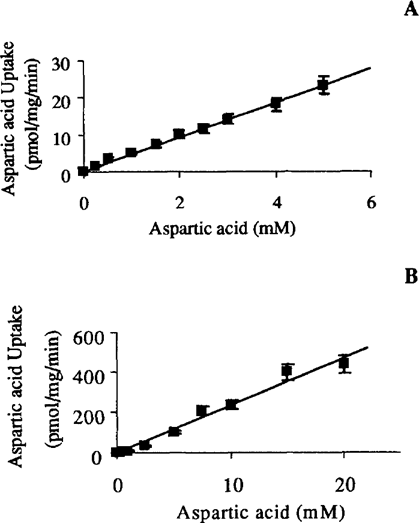
Uptake of radiolabeled aspartic acid by P. carinii. A. At pH 7.4. B. At pH 5.5. Uptake was directly related to extracellular substrate concentration, indicating simple diffusion as the transport mechanism. Uptake of this acidic amino acid was greater at acidic extracellular pH.
In contrast, the uptake by P. carinii of the neutral amino acids glutamine, leucine, and serine; the basic amino acid arginine; and the aromatic amino acid tyrosine exhibited Michaelis-Menten first order (saturation) kinetics, indicating that these compounds were transported by carrier-mediated mechanisms (Table 1).
| Substrate | Apparent Km | Vmax (pmol/ mg proteinAnin) | Reference |
|---|---|---|---|
| Neutral amino acids | 3 | ||
| Glutamine | 870–920 μM | 352–380 | |
| Leucine | 366–391 μM | 54–61 | |
| Serine | 268–273 μM | 513–520 | |
| Basic amino acid | 2 | ||
| Arginine | 23μM | 3 | |
| 4mM | 217 | ||
| Aromatic amino acid | 2 | ||
| Tyrosine | 284 μM | 93 | |
| Glucose | 5 mM | 3,560 | |
| 2-DOG | 67 μM | 237 | |
| 6 mM | 3,035 | ||
| SAM | 5 μM | 22,000 | 6 |
| 333 μM | |||
| Fatty acids | |||
| Oleic acid | 12μM | 7 | |
| Palmitic acid | 70 μM | 7 |
Kinetic parameters and competition by a variety of different amino acids for the transporter suggested that the neutral amino acids leucine, glutamine, and serine used the same carrier system for translocation into P. carinii. Glycine, leucine, and serine inhibited glutamine uptake, but asparagine, glutamic acid and arginine did not. Isoleucine, valine, threonine, glycine, and serine, inhibited leucine uptake, but asparagine, glutamine, phenylalanine, tyrosine, tryptophan, arginine, lysine, proline aspartic acid and glutamic acid did not. Leucine, glutamine, cysteine, and tyrosine inhibited serine uptake, but aspartic acid and arginine had no effect.
Exogenous glucose appeared to be required for maximal amino acid uptake by P. carinii, as observed in some other organisms such as Schizosaccharomyces pombe [8–10]. Galactose, fructose and mannose were less effective in stimulating serine uptake. The reciprocal was not observed, i.e., serine had no effect on the uptake of 2-DOG or glucose. Thus, serine and glucose uptake are not co-transported into P. carinii.
The uptake of serine, tyrosine and arginine transport did not require Na+ or K+; no inhibitory effects were observed in the presence of ouabain, or valinomycin. Furthermore, inhibitors of P. carinii ATP synthesis (iodoacetate, cyanide, azide, SHAM), drugs that cause dissipation of electrochemical ion gradients and transmembrane potentials (DCCD, CCCP, gramicidin) did not affect the uptake of the amino acids analyzed. Thus, we concluded that the mechanism of uptake of these neutral, basic and aromatic amino acids was facilitated diffusion. Cycloheximide, an inhibitor of protein synthesis in cytosolic ribosomes had no effect on serine uptake whereas chloramphenicol, an inhibitor of organellar protein synthesis inhibited serine uptake. The basis for chloramphenicol's effect on serine uptake by P. carinii is not understood.
The basic amino acid arginine appeared to enter the cell by a highaffinity carrier at low extracellular concentrations and also by a lowaffinity carrier. Both transporters appeared to be independent of that for the neutral amino acids. Lysine and leucine inhibited arginine uptake whereas glutamine, phenylalanine and aspartic acid did not.
It appears that P. carinii may have a separate carrier for the aromatic amino acids, which can also transport neutral amino acids (or two neutral amino acid transporters with preference for different groups of neutral amino acid species). Phenylalanine, tryptophan, leucine, valine, serine, and glutamine inhibited tyrosine uptake whereas proline, arginine, and aspartic acid did not. More experiments are needed to dissect the different transporters in greater detail. Analyses of other amino acids and the degree of competitive inhibition exerted by the full spectrum of different amino acids would clarify the functions of the different systems. After more detailed analyses have been performed we can then determine the relative importance of size, hydrophobicity and length of the side chain in the binding of different amino acids to different carrier systems and determine whether there are correlations with the rate of translocation across the cell surface membrane of this organism. The identification of several distinct amino acid transporter systems detected in these physiological and biochemical studies indicate that there are more than the one gene coding for amino acid carriers in addition to the amino acid permease gene identified in the P. carinii genome project EST database.
Glucose and 2-deoxyglucose uptake
Pneumocystis takes up glucose and 2-DOG by facilitated transport via two systems. The Q10 values for the high-affinity and low-affinity carriers were 2.12 and 2.09, respectively, which are indicative of carrier activities (enzyme reactions). Both transport systems were inhibited by mannose, galactose, fructose, galactosamine, glucosamine, and glucose but not by allose, 5-thio-glucose, xylose, glucose-6-phosphate and glucuronic acid.
Iodoacetate; SHAM, KCN, and 2, 4-DNP inhibited the high-affinity transporter, suggesting it required ATP. Ouabain, monensin, CCCP, and DCCD also inhibited 2-DOG uptake, and replacement of Na+ in the incubation medium with choline, inhibited 2-DOG uptake, suggesting that glucose uptake by P. carinii required Na+ and H+. The high-affinity system was also inhibited by the protein synthesis inhibitors cycloheximide and chloramphenicol. These results suggest that the mechanism of glucose uptake by the high-affinity system is active transport; transport against a chemical gradient needs to be established. Unlike the active 2-DOG high-affinity system, the properties of the low-affinity system indicate that it operates by facilitated diffusion mechanisms. This transporter probably functions only when exogenous glucose is present in high concentrations.
Unlike die human erythrocyte glucose transporter GLUT1, the P. carinii transporters recognized fructose and galactose and were relatively insensitive to cytochalasin B. Thus, these differences suggest that the P. carinii glucose transporter systems may be good targets for drug development.
The uptake rates of amino acids and glucose by P. carinii were dramatically slower than that reported for SAM [6] and fatty acids [7] (Table 1). In general, radiolabeled and fluorescent lipids are taken up by P. carinii much faster than water-soluble compounds [4]. Furthermore, uptake of glucose and the amino acids analyzed appears very slow in P. carinii compared to that observed in some other protists. For example, the high affinity leucine uptake system in S. pombe was reported to have a Km of only 45 μM, which is almost a magnitude of order lower than that in P. carinii. The slow uptake rates (low affnity, low capacity transporters) of amino acids by P. carinii suggests that the organism is capable of synthesizing de novo much of these compounds. The detection and sequencing of the P. carinii pentafunctional arom gene that encode for shikimic acid pathway enzymes [1] strongly suggests that P. carinii can at least synthesize de novo the aromatic amino acids.
ACKNOWLEDGEMENTS
We thank Michael Wyder for technical assistance. Supported in part by NIH grant RO1 AI29316.




