Distribution of DHPS Mutations Among ITS Subtypes of P. carinii f. sp. hominis
Sulfa drugs are widely used in the prophylaxis and treatment of Pneumocystis carinii pneumonia. Co-trimoxazole, the first-line agent, is a combination of sulfamethoxazole and trimethoprim but functions as sulfa monotherapy since P. carinii is apparently insensitive to trimethoprim. Dapsone, a sulfone, is a commonly used second-line agent. Both sulfamethoxazole and dapsone act by binding to dihydropteroate synthase (DHPS) (reviewed in references [2,19]) (Fig. 1).
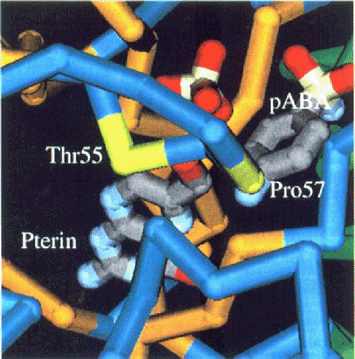
Amino acids in the E. coli DHPS homologous to mutated amino acids in the P. carinii DHPS.
Sulfa-resistant Pneumocystis carinii may be an emerging public health problem. However, since in vitro cultivation of human-derived organisms is not possible, molecular tools for determining resistance are needed. Genetic polymorphisms in the P. carinii DHPS were first reported in 1997 [11]. Two of the mutations, at positions 55 and 57, appear to be in the enzyme active site (Fig. 1, based on the E. coli structure [1]; MMDB id: 7741; PDB id: 1AJO). These mutations have been associated with sulfa exposure, prophylaxis break-through, or treatment failure [4,7,9–11,13–15,18].
These mutations appear to have occurred recently, and then to have spread fairly rapidly. In several studies, the mutations were infrequent in patient samples obtained before 1993, and then became more frequent as the sulfa drugs became used more widely in prophylaxis [4,9,10]. Also, the prevalence of these mutations varies geographically, with higher prevalences at sites with older epidemics [3,7,10].
How did the mutations evolve? There are two main possibilities. The mutations might have arisen only once and then spread as a result of selection on a population level. Alternatively, mutations could arise independently in each treated patient and increase as a result of selection within host. Obviously, intermediate scenarios (where the mutations arose independently but infrequently) are also possible.
MATERIALS AND METHODS
In order to understand how these mutations arose, we asked whether DHPS mutations were associated with individual strains of human-derived P. carinii. Strains were defined by sequencing the Internally Transcribed Spacer (ITS) regions. Human-derived P. carinii contain >15 ITS1 types (A-O) and >14 ITS2 types (a-n). More than 50 different combinations have been identified [5,8,12]. We also assessed the phylogenetic relatedness of strains with and without DHPS mutations. If strains with the mutation comprise a set of closest relatives that would also be consistent with a single origin of resistance. However, we did not anticipate that any association would be perfect, since ITS and DHPS genes are probably unlinked and there could be horizontal genetic transfer.
We obtained ITS and DHPS genotyping data from 57 patient isolates retrospectively and prospectively. These patients were from Indiana, Atlanta, San Francisco, Los Angeles, and Seattle and were a subset of patients used in previous studies [3,7,9–11]. We then selected all 37 isolates with monoclonal infections (only single ITS type and DHPS type).
RESULTS AND DISCUSSION
Characteristics of 37 monoclonal patient isolates are shown in Table 1. There were 22 wild types, 14 double mutants and 1 single mutant (position 55). Mutations were more common in Atlanta and the West Coast than Indiana, and more common in samples obtained more recently, but these differences were not statistically significant.
| Characleristic | No. | Mutant (%) |
|---|---|---|
| Site | ||
| Indiana | 17 | 3 (18) |
| Atlanta | 15 | 9 (60) SFILAlSeattle |
| SF/LA/Seattle | 5 | 3 (60) |
| Year | ||
| 92–97 | 14 | 3 (21) |
| 98–99 | 23 | 12 (52) |
One ITS type, Eg, was common in all 3 sites (Table 2). Three other types, Ea, Eb, and Ne, were found in two of the 3 sites. The other 6 ITS types were only found in single sites.
The distribution of DHPS mutations among ITS types is shown in Figure 2. Mutations are very common in some ITS types (such as Ne), but not found in others.
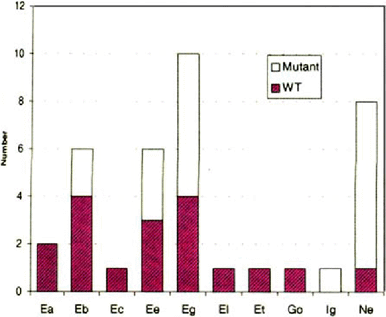
Distribution of mutant DHPS genotypes among ITS types.
A phylogenetic hypothesis for ITS types of P. carinii, along with the association of DHPS mutations by type is shown in Fig. 3. Consensus sequences for ITSl and ITS2 were combined in the same manner as found in the isolates in our study. (Genbank accession numbers are: ITSl Type C: AF013808, E: AF013810, G: AF013812,[:AF013814, M: AF013818, N: AF013819; ITS2 Type a: AF013821, b: AF013822, c: AF013823, e: AF013825, g: AF013827, o: AF013835, t: AF013840.) We also included two P. carinii sequences from Macaca mulatta [6] as outgroups (ITS1 Type A: AF288827, D: AF288834; 1TS2 Type d: AF288843, f: AF288846). Sequences were aligned in ClustalW [17] with gap extension penalties set to 7.5.
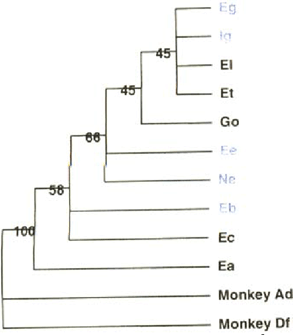
Evolutionary relationships between isolates used in this study. ITS types in blue contain isolates with DHPS mutations.
A phylogenetic hypothesis was constructed using parsimony analysis as implemented in PAUP*4.0b4a [16]. With gaps considered as a fifth character state, 171 characters were constant and 92 were parsimony informative. A search (assessing all possible tree topologies) resulted in a single most parsimonious tree 262 steps in length (Fig. 3), which we rooted with the P. carinii sequences from M. mulatta. Values to the left of the nodes within the tree indicate the relative support for that particular clade based on 1000 bootstrap resamplings of the sequence data. P. cariii strains with and without DHPS mutations appear well mixed in the phylogeny.
We then asked post-hoc, whether certain groups of ITS types contained statistically higher numbers of DHPS mutants than other ITS types using the Fishers' exact test (Table 3). Among isolates from Indiana, Ee isolates were significantly more likely to be DHPS mutants than all others. Among all isolates, Ne, Eg and Ee isolates were more likely to contain DHPS mutants than all others. This is not an artifact associated with date of acquisition, since Ee and Eg are among the oldest of the isolates collected. Thus, our data are consistent with the possibility that DHPS mutations arose infrequently but on more than one occasion.
| ITS type | DHPS mutam | WT | P value |
|---|---|---|---|
| Indiana only | 0.03 | ||
| Ee | 3 | 3 | |
| Others | 0 | 11 | |
| All samples | 0.003 | ||
| Ne, Eg or Ee | 16 | 8 | |
| Others | 3 | 10 |
ACKNOWLEDGEMENTS
We would like to thank Marilyn Bartlett, Jim Smith, Wendy Armstrong, Carlos del Rio, Jeffrey Duchin, James Richardson, David Rimland, Paul Simon for access to patient samples. This work was supported by NIH grants RO1 AI 46966 and T32 HG00040.





