Three New Karyotype Forms of Pneumocystis carinii f. sp. carinii Identified by Contoured Clamped Homogeneous Electrical Field (CHEF) Electrophoresis
Two distinct populations of Pneumocystis have been identified in rats obtained from commercial vendors. These two populations have been found to co-exist within an individual rat lung. Pneumocystis carinii f. sp. carinii is used to designate the population previously called ‘prototype’ and Pneumocystis carinii f. sp. ratti is used to designate ‘variant’[1]. These two populations are identified by distinct karyotypic profiles and fidelity and divergence from the small ribosomal subunit gene sequences originally reported for rat derived Pneumocystis [2,5].
Eight distinct banding patterns (forms) produced by pulse field gel electrophoresis have been documented for P.c. carinii illustrating this population has more chromosomal length polymorphisms (CLPS) than that of P.c. ratii, which has only one karyotypic profile reported. These banding patterns have been shown to be stable and reproducible over time. Low levels of genetic polymorphisms characterize the P.c. carinii karyotype forms.
In the present study, a colony from Charles River Laboratories (Wilmington, Mass.), and Hill Top Laboratories (Scottsdale, Pa.); 3 colonies from Harlan Industries (Indianapolis, Ind.); and 3 colonies from Taconic Farms (Germantown, NY) were surveyed in the search for a Pneumocystis-free colony. Form 1 was detected in the Charles River (Raleigh 12) colony, form 7 was harbored in the Harlan and Hill Top rats and form 8 was found at Taconic (MBU 4). There were 4 new forms detected (10, 11, 12, and 13) in the Taconic facilities (IBU 6, IBU 18, and MBU 4), as well as another form found in 1 rat (9). The requirement for the designation of a new karyotype form was defined as the presence of the same banding pattern found in at least 3 different rats. Unlike previous surveys, 2–3 of the new forms could be found in a single colony. Forms 10, 11, and 12 also displayed the ability to develop a mixed infection in a single rat lung heretofore only observed with P.c. carinii and P.c. ratti. These data illustrate that P.c. carinii displays even more extensive chromosomal length polymorphisms than previously thought.
MATERIALS AND METHODS
Induction of experimental P.carinii f. sp. carinii infections
Various strains of male rats were received in filtered containers at 125 to 150 g from various commercial rat vendors. Upon receipt, the rats were immediately placed under barrier isolation consisting of sterile polycarbonate shoebox cages with sterile bedding; the cages had been fitted with microisolator tops placed on stainless steel racks within a horizontal flow hood. To reduce the occurrence of infection with other microbial pathogens, the rats received irradiated food (Tek Lab Irradiated Chow; Harlan Industries, Indianapolis, Ind.) and autoclaved water. After 7 days of acclimation, the rats were started on a regimen of immunosuppression consisting of weekly injections of methylprednisolone acetate (Depo-Medrol; The Upjohn Co., Kalamazoo, Mich.) at 4 mg/kg/week. The duration of the immunosuppression lasted 10–12 weeks.
Preparation of P. carinii f.sp. carinii organisms from rats
Rats were sacrificed by carbon dioxide inhalation, and the lungs were removed and processed for Clamped Contour Homogeneous Electric Field (CHEF) electrophoresis as previously described [1]. Briefly, the lungs from individual rats were homogenized with a laboratory blender (Stomacher 80; Tekmar, Inc., Cincinnati, Ohio). Large particles were removed by sieving the material through sterile gauze, and the homogenates were treated with aqueous ammonium chloride (0.85%) to lyse the erythrocytes and some host cells. Host cell numbers were further reduced by at least two passes through 10-μm-pore-size filters (Mitex; Millipore Corp., Bedford Mass.). The organisms were then treated with DNase I (Boehringer Mannheim Biochemicals, Indianapolis, Lid.) at 10 μg/ml in a solution of 150 mM NaCl-10 mM MgCl2–10 mM Tris at pH 7.2 for 30 min. at 37°C to digest extracellular DNA and then washed once in 250 mM EDTA and once in 125 mM EDTA. Organisms were embedded in low-melting-point agarose (Boehringer Mannheim) at a final concentration of 0.8% in disposable plug molds (Bio-Rad, Hercules, Calif.) or in disposable spectrophotometric cuvettes (Fisher Scientific), depending on the organism densities. Gel embedded organisms were digested with 0.25 mg of proteinase K (Boehringer Mannheim) per ml in a solution of 1% N-lauroylsarcosine (Sigma)—0.45 M EDTA—0.01 M Tris in a 55°C water bath for 24–48 hours. Digested samples were stored at 4°C in 0.5 M EDTA.
CHEF conditions. Gels for contour-clamped homogeneous electrical-field (CHEF) electrophoresis contained 1% FMC SeaKem GTG Agarose (SeaKem, Rockland, Maine) prepared in 0.5 X TBE (45mM Tris HCl, 45 mM boric acid, 1.25 mM EDTA) for a total volume of 190 ml and final dimensions of 14 by 21 cm. Electrophoresis was performed using a Bio-Rad CHEF DRII or CHEF DRIII apparatus. Gels were run for 105 to 115 hours at 13°C in 0.5 X TBE at with 4.1 V/cm with a 50-s initial pulse that was gradually increased to 100-s [1]. Chromosome-sized DNA were visualized with the nucleic acid stain SYBR-Gold (Molecular Probes, Inc., Eugene, Oreg.)
Pneumocystis DNA extractions
P.c. carinii DNA was extracted from low melt agarose plugs using the Eppendorf Phase Lock Gel Protocol for the recovery of DNA from LMP agarose (Eppendorf, Hamburg, Germany).
PCR Conditions
The PCR was performed on the DNA extractions described above. Primers directed to the mitochondrial large subunit rRNA (mtlsu) were used to amplify at least 3 representatives of each of the P. carinii f. sp. carinii karyotype forms 10, 11, 12, and 13 whereas only one sample of form 9 was available for analysis. Template (1.5 μl) and primers (200 nmols) were added to each reaction. JumpStart RED Taq Ready Mix (Sigma, St. Louis, Miss.) was used with a hot start of 94°C for 2 min. PCR conditions are listed in Table 1 below and primer sequences are listed in Table 2.
| Primer Set | Product | Denaturing | Annealing | Extension | cycles |
|---|---|---|---|---|---|
| globin 1&2 | 400 bp | 94°C/30 s | 50°C/30 s | 72°C/30 s | 30 |
| pAZ 102 H&E | 346 bp | 94°C/30 s | 54°C/30 s | 72°C/30 s | 35 |
| RC 1&2 | 137 bp | 94°C/30 s | 54°C/30 s | 72°C/30 s | 40 |
| RR 1&2 | 251 bp | 94°C/30 s | 54°C/30 s | 72°C/30 s | 40 |
| Primer | Sequence (5′–3′) |
|---|---|
| Globinl | GGT GCA CCT AAC TGA TG |
| Globin2 | GCT TGT CAC AGT GGA GTT CAC |
| pAZ 102 H | GTG TAG GTT GCA AAG TAG TC |
| pAZ l02 E | GAT GGC TGT TTC CAA GCC CA |
| RC 1 | TTT TGG TAG ATG ACT TGT TAT T |
| RC 2 | AGT CTG ATA ACT CAT CAT ATA T |
| RR 1 | GTA GAT AGC TTA ATA AGG ATG |
| RR 2 | TTC TTG ACT GTC TAT GAA GT |
The globin primers from O'Leary et al. [3] were used for the detection of rat DNA. The pAZ 102, H&E primers were used to amplify the large subunit of rat Pneumocystis carinii mitochondrial rRNA as reported by Wakefield et al. [6]. Pneumocystis carinii f. sp. carinii -specific RC primers and Pneumocystis carinii f. sp. ratti RR primers were used to identify the identities of the P.c. carinii populations and first reported by Palmer et al. [4]. Amplicons were visualized by ethidium bromide staining of 1% agarose gels.
Nucleotide sequencing
P.c. carinii DNA was purified using the QIAquick PCR purification kit (Qiagen, Valencia, CA) and sent to Genomatics (Norwood, OH) for sequencing with an Applied Biosystems 373 sequencer. Sequences were aligned using DNAman software (Lynnon Biosoft, Montreal, Quebec).
RESULTS AND DISCUSSION
Eight colonies from four commercial rat vendors were surveyed (Table 3). Form 1 was found only in the Charles River facility. Form 7 was found in both Harlan and Hill Top facilities. Forms 8, 9, 10, 11, 12, and 13 were found exclusively in colonies at the Taconic facility. MBU 4 and IBU 18 had various forms within a single colony. Mixed infections of P.c. carinii forms were also detected in the MBU 4 and IBU 18 colonies. CHEF separations of all forms of Pneumocystis carinii f. sp. carinii, Pneumocystis carinii f. sp. ratti, and some mixed infections are shown in Fig. 1.
| VENDOR | COLONY | STRAIN | FORMS PRESEN T | INFECTED RATS/TOTAL |
|---|---|---|---|---|
| CHARLES RIVER | RALEIGH 12 | CD | 1 | 2/12 |
| HARLAN | NA | HOLTZMA N | 7 | 4/12 |
| NA | LONG EVANS BS | 7 | 13/19 | |
| NA | SD | 7 | 4/12 | |
| HILLTOP | HBM176/E MERY | SD | 7 | 1/24 |
| TACONIC | MBU4 | SD | 8,10,11,12 | 32/48 |
| IBU6 | F344 | 13 | 4/12 | |
| IBU18 | SD | 9*, 10,11,12 | 17/36 |
- *Form 9 was found in 1 rat. NA—not available. BS—Blue Spruce. SD—Sprague Dawley.
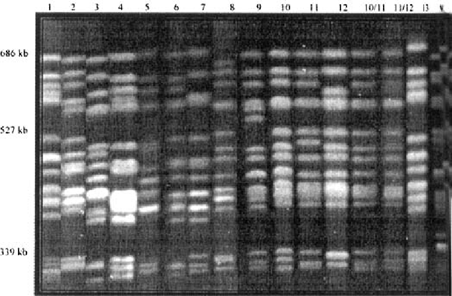
CHEF separations of Pneumocystis carinii f. sp. carinii forms 1–13 and mixed infections.
PCR products from putative new P.c. carinii forms were the expected size for each set of primers used. An amplicon of each new form was sent for sequencing. All of the samples showed 100% identity with the mtLSU sequence of forms 1, 3–8, proving these organisms were P. carinii f. sp. carinii in nature (Fig. 2). Surprisingly, no evidence of P.carinii f. sp. ratti was found by karyotyping.
PCR Results from new forms using pAZ 102, H&E primers.
Pneumocystis carinii f. sp. carinii was the prevalent species in the commercial rat vendor colonies surveyed and displayed extensive chromosomal length polymorphisms that were resolved within the same size range. These surveys also showed the widespread occurrence of P. carinii in commercial rat colonies.
ACKNOWLEDGMENTS
Supported by NTH grants RO1-AI-44651 and RO1-AI-29839.




