Pneumocystis carinii Infection in Red-Bellied Tamarins and Cynomolgus Monkeys, and the Characterization of the Mitochondrial Large Subunit Ribosomal RNA Gene of Pneumocystis carinii
Pneumocystis carinii (P. carinii) is a causative agent of opportunistic infection, is widely distributed among a variety of animal species including human beings, and causes fatal pneumocystosis in immunocompromised hosts like those with AIDS [8]. Cases of fatal and systemic pneumocystosis have been reported in rhesus monkeys (Macaca mulatta) infected with simian immunodeficiency virus and activation of latent P. carinii infection in these monkeys has also been observed [1,7]. The reported data imply that this is a common opportunistic infection in monkeys. P. carinii infection has been reported in various new and old world monkey species [3–5], but there have been no reports of infection in red-bellied tamarins (Saguinus labiatus). Recently, we had an opportunity to examine P. carinee-infected red-bellied tamarins and cynomolgus monkeys and found a high incidence of P. carinii infection in these animals by histopathologic examination. We also examined the mitochondrial large subunit ribosomal RNA gene (mt LSU rRNA) of P. carinii derived from these two species and then compared their sequences with those of P. carinii derived from other mammalian hosts.
MATERIALS AND METHODS
The 10 red-bellied tamarins and 66 cynomolgus monkeys used in the study were born in a laboratory breeding colony in the Tsukuba Primate Center (TPC) and were maintained there individually in isolated cages until death. Postmortem examinations were performed on all monkeys within 12 hours of their natural death. Their lungs were removed and fixed with 10% neutral buffered formalin, and paraffin sections were made for histopathologic examinations. Hematoxylin-Eosin (HE), Grocott's, and toluidine blue O (TBO) staining were performed as described previously [2]. Immunostaining with avidin-biotin complex (ABC) using either rabbit sera immunized against P. carinii derived from mouse, rat, and simian hosts or a mouse monoclonal antibody against human P. carinii (Dako, Carpinteria, CA), was conducted to detect P. carinii cysts in the lungs. P. carinii DNA was extracted from the lungs of P. carinii positive tamarin and cynomolgus monkey by a combination of proteinase K digestion and phenol/chloroform extraction. Other P. carinii DNA was also extracted from the fresh lungs obtained from mouse, rat, rhesus monkey, human or paraffin-embedded lung tissues obtained from cat, dog, or horse having spontaneously severe pulmonary pneumocystosis, using the same method. The oligonucleotide primer pairpAZ102-H and pAZ102-E, designed to the P. carinii gene encoding the mt LSU rRNA, were used for PCR according to previously reported methods [9]. DNA bands amplified by PCR were cut from agarose gels, extracted with phenol/chloroform and cloned using the TA Cloning Kit (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. DNA sequencing was performed using an Automated DNA Sequencer. Phylogenetic analysis of the P. carinii mt LSU rRNA gene to assess phylogenetic distance was performed using CLUSTAL W. The alignment was bootstrapped 1000 times, the maximum likelihood distance matrix data were collected, and a consensus neighbor-joining tree was generated.
RESULTS AND DISCUSSION
At autopsy, lungs showed almost normal findings, and P. carinii cysts were detected in 6 of 10 tamarins and 1 of the 66 cynomolgus monkeys upon histopathologic examination, although there were a low number of cysts. The 3–4 μm sized cysts stained with ABC were attached individually to the alveolar surface or were in alveolar spaces without any infiltration into the inflammatory cells. The cysts were barely detectable by HE staining, but were stained as black ovum or round shaped objects with Grocott's or were stained purple with TBO, and were colored brown with ABC staining using different sources of anti-P. carinii sera. Except for these findings, no histopathologic changes were found in the lungs, and the cysts observed by histopathologic examination in this study were morphologically identical to P. carinii cysts derived from other hosts. We used the ABC method to detect P.carinii cysts using antisera to various P. carinii derived from different hosts. Since the cysts of the tamarin were stained by these antisera, it can be assumed that the cysts share common antigens with P. carinii from other organisms [6]. The findings obtained from the present studies suggest that P. carinii may be an important pathogen in this species.
P. carinii strains have been isolated from the lungs of a variety of mammalian hosts, and all share a common morphology although immunological and genetic studies indicate that these organisms are not identical. To examine the genetic diversity of this pathogen, we sequenced PCR products of mt LSU rRNA of P. carinii derived from tamarin and cynomolgus monkey-P, carinii; fragment lengths were 315 bp and 307 bp, respectively. Genetic diversity in P. carinii DNA was found in both types of monkeys, indicating that P. carinii derived from different hosts are genetically distinct. Furthermore, the tamarin and cynomolgus monkey-P. carinii DNA sequences were aligned with those derived from 6 different mammalian hosts. Tamarin and cynomolgus monkey-P.carinii sequences contained regions with significant homology to P. carinii sequences isolated from other hosts, in addition to regions that were divergent. Phylogenic trees were constructed from each alignment using the neighbor joining method, in the form of a bootstrapped unrooted tree. The results of the analysis suggested that cynomolgus monkey-P, carinii is closer to tamarin-P. carinii than that of rhesus monkey, although cynomolgus and rhesus monkeys are taxonomically in the same group of Macaca monkeys (Fig. 1). On the other hand, the rhesus monkey-P. carinii is close to human-P. carinii, and then, rat- and mouse-P. carinii. These results show that tamarin- and cynomolgus monkey-P. carinii seem to be different from groups of other P. carinii, and that each P.carinii has its own type of organism with a specific mt LSU rRNA gene, although these data are still preliminary. More specimens will be sequenced in the future to reconfirm the results obtained here and we will also analyze other genes of P. carinii to examine the differences among P. carinii derived from different hosts.
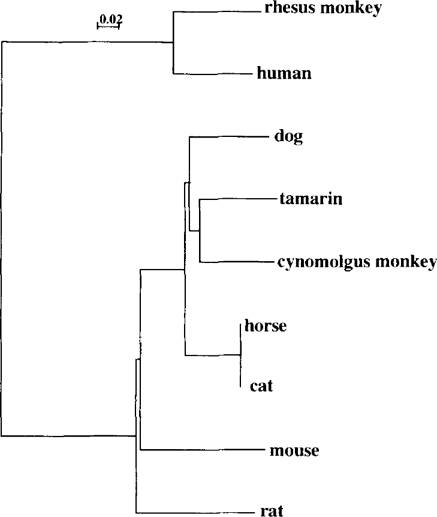
Phylogenetic tree inferred from mt LSU rRNA sequences corresponding to different Pneumocystis organisms.




