Microarray-based Expression Analysis of Human Epithelial Cell Response to Cryptosporidium parvum Infection
The apicomplexan Cryptosporidium parvum infects primarily the epithelial cells of the small intestine of mammals and causes gastrointestinal disease that is characterized by acute watery diarrhea. The severity of the disease is largely dependent on the immune status of the host and could become persistent and life threatening in patients with deficient immune system. There is currently no effective therapy to treat the disease and efforts to develop novel therapeutical strategies are hampered by the limited understanding of the mechanism by which C. parvum causes disease, especially the host-parasite interaction [1]. As of today, little is known about the responses of host epithelial cells to C. parvum infection, such as biochemical pathways that are affected or genes that are specifically regulated.
In the past few years, the development of DNA microarray technology has allowed mRNA expression to be assessed on a global scale [2]. By measuring transcription levels of genes in an organism under various conditions, differential expression profiles for tens of thousands of genes could be assessed in parallel. Systematic application of this approach has been widely used for the identification of candidate genes involved in a variety of processes. This study was designed to develop and apply a human cDNA microarray in investigating human epithelial cell response to cryptosporidial infection and to identify host genes regulated during intracellular C. parvum development.
MATERIALS AND METHODS
Preparation and immobilization of human cDNA probes
Human cDNA clones were obtained from Research Genetics (Huntsville, AL) and cDNA inserts were amplified, in 100-μl reactions and in 96-well format, from overnight cultures using vector-specific flanking primers. Following amplification, a small portion (∼4 μl) of each reaction was analyzed on agarose gel to determine the efficiency of amplification, the remaining PCR reactions were transferred into MultiScreen glassfiber filter plates (Millipore Corporation, Bedford, MA), and PCR products were purified using a MultiScreen vacuum manifold (Millipore). The filter-bound DNA was washed twice with distilled water, dissolved in 3 × SSC, and pipeted into 384-well plates (Nalge Nunc International, Rochester, NY). The amplified cDNA inserts were spotted onto poly-1-lysine coated glass slides using a MicroGrid II spotting machine and MicroSpot 2500 pins (BioRobotic Inc., Woburn, MA). A set of the cDNA inserts was spotted as duplicates on each slide and the distance between spots was 250 μm. Prior to hybridization, the spotted slides were rehydrated and the slide surface was blocked with succinic anhydride following standard protocols [4].
Preparation and labeling of cDNA targets
Sterilized C. parvum oocysts were used to infect human adenocarcinoma (HCT-8) cells (American Type Culture Collection, Manassas, VA) at a concentration of one oocyst per cell, as previously described [3,5]. Total RNA was extracted from mock-infected or C. parvum-infected cells by using Trizol reagents (GIBCO Life Technologies, Gaithersburg, MD) and mRNA was isolated by using oligo-dT-cellulose column (Amersham Pharmacia Biotech, Piscataway, NJ). First-stranded cDNA was synthesized using the SuerScript cDNA synthesis system (GIBCO) and labeled by indirect incorporation of fluorescent dye Cy3 or Cy5 as described below.
For reverse transcription, 2.0 μg of mRNA was first combined with 4.0 μg of oligodT primer (GIBCO) and 2.0 μg of random hexamer (GIBCO) to a volume of 14.5μl, heated to 65°C for 10 min, and cooled on ice. The remaining components (15.5 μl) were then added to obtain the following reaction conditions (in 30 μl total reaction): 1×1st strand buffer, 100 mM DTT, 500 μM dATP, 500 μM dCTP, 500 μM dGTP, 300 μM dTTP, 200 μM aminoallyl-dUTP (Sigma Diagnostics Inc., St. Louis, MO), and 3.8 μl of Superscript II reverse transcriptase (GIBCO). Following a 2-h incubation at 42°C, the reaction was stopped by adding 10 μl of 500 mM EDTA, the RNA was hydrolyzed by adding 10 μl of 1.0 N NaOH and incubating at 65°C for 15 min, and the reaction was neutralized by adding 25 μl of 1.0 M Tris-HCl (pH 7.4). The cDNA was then concentrated using a Micron 30 concentrator (Millipore), vacuum-dried, and stored at -20°C. For Cy3/Cy5 coupling, the cDNA was resuspended in 4.5 μl of H2o and mixed with Cy3 or Cy5 (Amersham) that was resuspended in 4.5 μl of 0.1 M sodium bicarbonate (pH 9.6). Following 1-h incubation at room temperature, uncoupled Cy3/Cy5 was quenched by adding 4.5 μl of 4.0 M hydroxylamine (Sigma) and incubating at room temperature for 15 min. Two labeled cDNAs were then combined and separated from unincorporated & quenched dyes using the QIAquick PCR purification kit (QIAGEN Inc., Valencia, CA). The volume of eluted cDNA target was decreased to ∼15 μl by vacuum drying.
Array hybridization and data analysis
The labeled cDNA targets were combined with 4.5 μl of 20 × SSC, 0.68 μl of 10% SDS, and 2.3 μl of 10 mg/ml poly A (Sigma) to a final volume of 30 μl. The mixture was boiled for 5 min, cooled at room temperature for 5 min, and applied to the human cDNA array slide that was placed in an array hybridization chamber (TeleChem International Inc., Sunnyvale, CA). A coverslip was added and the hybridization chamber was sealed and immediately transferred into a 65°C waterbath. Following overnight incubation, the array was washed in 1 × SSC-0.1% SDS for 5 min and then in 0.1 × SSC for 5 min, and dried by spinning at 500 g for 5 min. It was scanned in a ScannArray 5000 (GSI Lumonics, Watertown, MA) and analyzed using the QuantArray software (GSI Lumonics). The Cy3 and Cy5 signal intensities relative to each array were standardized by comparing the mean signal intensities of all spots and the relative expression of a gene in two samples were determined by the signal intensity of the spots.
RESULTS AND DISCUSSIONS
From 5,184 human clones, 4,918 (94.87%) were successful amplified and spotted. To identify genes involved in human epithelial cell response to cryptosporidial infection, we hybridized the array with Cy3-labeled cDNA targets prepared from mock-infected HCT-8 cells and Cy5-labeled cDNAs from C. parvum-infected cells at 24 h postinfection. Fig. 1 represents sections of the human cDNA array after hybridization and composition of the images. The relative intensity of Cy3 (red) to that of Cy5 (green) correlated to the relative abundance level of that gene in two samples. Therefore, genes highly expressed in mock-infected cells are red and genes highly expressed in infected cells are green, indicating those genes are down- and up-regulated, respectively. Genes that are expressed at similar levels in both samples are yellow or brown, whereas genes that are not expressed in either sample are represented by black spots.
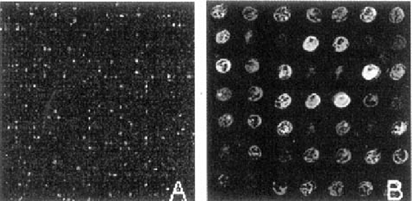
Sections of the human cDNA array. cDNA targets from mock-infected and C. parvum-infected (24 h post-infection) HCT-8 cells were labeled with Cy3 and Cy5, respectively.
As it is shown in figure 1B, infection of human epithelial cells with C. parvum resulted in differential regulation of gene expression. We're currently in the process of repeating the experiments and identify genes that are consistently up- or down- regulated upon infection. RT-PCR and Northern blot analysis will be conducted to confirm the microarray findings. Further characterization of these genes will provide important clues for understanding the biochemical changes that occur within host cells in response to C. parvum infection and provide insights of the disease progression.
ACKNOWLEDGMENTS
This research is supported partly by the National Institute of Health (USA). We thank Mr. Alan Wang, Mr. Mike Paustian, and Ms. Cheryl Lancto for technical assistance.




