Multilocus Genotyping of Cryptosporidium Isolates from Human HIV-Infected and Animal Hosts
The genus Cryptosporidium comprises parasites of several species that infect mammals and other vertebrates. Molecular studies have shown that Cryptosporidium parvum, the most common species infecting man, comprises the “human” genotype (H) found exclusively in humans and the “cattle” genotype (C) found in both humans and livestock [8]. Other genotypes, until now considered to be host specific, have also been found in mice, pigs, ferrets and marsupials [5,14]. Recently, PCR-RFLP and DNA sequence analysis have also identified C. felis, C. meleagridis and a Cryptosporidium“dog type” in HIV-infected patients as well as in immunocompetent persons [4,6,7,9,12].
In this study we characterized Cryptosporidium isolates from AIDS patients and animals from Portugal, by a PCR-RFLP multilocus analysis.
MATERIALS AND METHODS
Cryptosporidium isolates from 25 HIV-infected patients with diarrhoea and from 20 animals –12 calves, two lambs, five wild ruminants from the Lisbon Zoo and one cat—were studied.
Oocysts were concentrated by a modified water-ether sedimentation method [1]. DNA was isolated from concentrated oocysts by a MiniBeadBeaters/Silica method [4].
PCR amplification and subsequent RFLP analysis with the restriction cndonuclease Rsal of the thrombospondin-related adhesive protein of Cryptosporldiwn-l (TRAP-C1) and Cryptosporidium oocyst wall protein (COWP) gene fragments were performed as described previously [1], A two-step nested PCR of a 18S small-subunit (SSU) rRNA gene fragment and restriction fragment analysis with the endonucleases VspI and SspI were carried out as previously described [13]. A fragment of the dihydrofolate reductase (DHFR) gene [3] was amplified by a modified nested PCR protocol and digested with the endonuclease BpuAl as previously described (M. Alves, accepted for publication),
RESULTS AND DISCUSSION
The PCR-RFLP analysis of the isolates from HIV-infected patients showed that eight were infected with the “H”C. parvum genotype, 14 with the “C”C. parvum genotype, one with C. fells, one with C. meleagridis and another with a non-C. parvum isolate. All the animal isolates presented the “C”C. parvum genotype, with the exception of a C. fells and a non-C. parvum isolate found, respectively, in a cat and in a calf (Table 1).
| Human Isolates | Animal Isolates | |
|---|---|---|
| C. parvum | 8 | — |
| “H” genotype | ||
| C. parvum | 14 | 18 (ruminants) |
| “C” genotype | ||
| C. felts | 1 | 1 (cat) |
| C. meleagridis | 1 | — |
| Non-C. parvum | 1 | 1 (calf) |
The C. fells and the non-C. parvum isolates were differentiated from the other species by digestion of the SSU-rRNA amplicon with the endonuclease Sspl (Fig. 1B). When amplified with the DHFR primers, one isolate (CT.14, Fig. 1A) originated an amplicon of unexpected size which digestion with the BpuA1 endonuclease showed an absence of a recognition site to that restriction enzyme. The species differentiation of that isolate was done by PCR-RFLP analysis of the COWP and SSU-rRNA genes and it was identified as C. meleagridis.
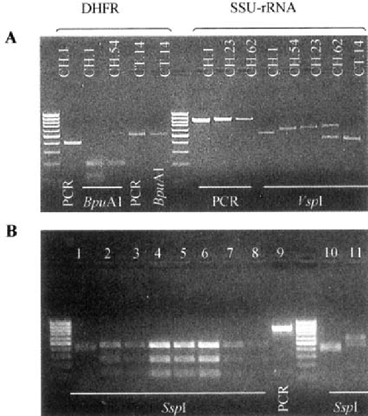
Gel electrophoresis of PCR and digestion products of Cryptosporidium isolates. A. CH.l: C. parvum“H” genotype; CH.54: C. parvum“C” genotype; CT.14: C. meleagridis; CH.23: C. felis; CH.62: non-C. parvum isolate. B. Lane 1: C. felis; lanes 2–8: C. parvum; lane 9: PCR product of a C. parvum isolate; lane 10: non-C. parvum isolate from an AIDS patient (CH.62); lane 11: non-C. parvum isolate from a calf. Marker: 100 bp ladder.
From the four protocols used in this multilocus genotyping study, the nested PCR-RFLP analysis of the genes DHFR and SSU-rRNA were the most sensitive techniques for the amplification of parasite gDNA, although the former have shown a lower resolution as a genotyping and species differentiation tool.
In the Portuguese HIV-infected population studied, we found a higher number of patients infected with the “C”C. parvum genotype. Some studies have also reported a predominance of the “C”C. parvum genotype in HIV-infected patients from Switzerland [6] whereas in others a larger number of the “H”C. parvum genotype isolates was found in AIDS patients from the United States [9–11]. However, the significance of these results is limited by the small number of samples involved. All the ruminants were infected with the “C”C. parvum genotype, with the exception of a non-C. parvum isolate found in a calf.
Additional studies should be done in order to identify the two non-C. parvum isolates with amplicon sizes and RFLP patterns different from those of Cryptosporidium parasites already described (one from an AIDS patient and other from a calf).
More discriminatory genotyping tools and a larger number of samples are needed in order to evaluate the public health significance of various Cryptosporidium species and genotypes in Portugal and to enhance our understanding about the transmission of cryptosporidiosis.
AKNOWLEDGEMENTS
This work was supported in part by the Biomed 2 Project BMH4 CT97 2557 from the European Commission




