Linkage Analysis of Alcohol Dependence Symptoms in the Community
Abstract
Background: We have previously identified suggestive linkage for alcohol consumption in a community-based sample of Australian adults. In this companion paper, we explore the strength of genetic linkage signals for alcohol dependence symptoms.
Methods: An alcohol dependence symptom score, based on DSM-IIIR and DSM-IV criteria, was examined. Twins and their nontwin siblings (1,654 males, 2,518 females), aged 21 to 81 years, were interviewed, with 803 individuals interviewed on 2 occasions, approximately 10 years apart. Linkage analyses were conducted on datasets compiled to maximize data collected at either the younger or the older age. In addition, linkage was compared between full samples and truncated samples that excluded the lightest drinkers (approximately 10% of the sample).
Results: Suggestive peaks on chromosome 5p (LODs >2.2) were found in a region previously identified in alcohol linkage studies using clinical populations. Linkage signal strength was found to vary between full and truncated samples and when samples differed only on the collection age for a sample subset.
Conclusions: The results support the finding that large community samples can be informative in the study of alcohol-related traits.
Alcohol dependence is associated with significant economic burden, both directly and via its comorbidity with other substance use disorders and psychopathology. Data collected for the Australian National Survey of Mental Health and Wellbeing in 1997 showed that 4.1% of those surveyed met the criteria for DSM-IV alcohol dependence (19% meeting at least 1 DSM-IV alcohol abuse or dependence criterion) in the preceding 12 months (2006). Findings are similar in the United States, with prevalence for 12-month and lifetime DSM-IV alcohol dependence being 3.8 and 12.5% (Hasin et al., 2007).
It is now known that genetic factors influence variation in alcohol dependence, with heritability estimates reported to range 50 to 70% (Agrawal and Lynskey, 2008; Heath et al., 1997; Prescott and Kendler, 1995, 1999). Over the last 2 decades, several linkage scans have been undertaken to locate and identify the specific genes that contribute to alcohol dependence. Among these efforts are the Collaborative Study on Genetics of Alcoholism (COGA) (Begleiter et al., 1995; Reich et al., 1998) and the Irish Affected Sib-Pair Study of Alcohol Dependence (IASPSAD) (Prescott et al., 2005, 2006), which have used linkage methods to identify loci on several chromosomes. These studies have focussed on patients with a family history of drinking problems.
In the current study we utilize data collected from a community sample of over 4,000 Australian adults. In line with our previous study (Hansell et al., 2009), which found suggestive linkage for alcohol consumption in the same hetereogeneous sample, the primary aim of the present study was to examine the viability of using linkage methods on alcohol dependence data collected from a community sample. In addition, we investigated the effects of excluding very light drinkers (approximately 10% of the distribution of consumption). This exclusion was undertaken to provide a focus on genetic factors predisposing to problem drinking. Low consumption may reflect a mixture of both biological causes (which would include protective genes such as the aldehyde dehydrogenase gene ALDH2 (Luczak et al., 2006) and which may be evident in analyses of the full distribution) and social factors (such as church attendance (Strawbridge et al., 1997). Third, as data were collected on 2 occasions (10 years apart on average) for approximately 19% of our sample, we examined the effect of including data collected at either the younger or the older age. Lifetime dependence symptomatology can vary with age, either due to increasing symptoms with age, or due to incorrect recall of symptoms that were apparent at the earlier, but not the later, age.
Materials and Methods
Sample
Data were obtained from 3 phases of telephone interviews conducted between 1992 and 2005, as described previously (Hansell et al., 2009). Interviews were adapted from the Semi-Structured Assessment for the Genetics of Alcoholism (SSAGA, Bucholz et al., 1994). Lifetime abstainers (i.e., individuals who had never tried alcohol −2% of those interviewed) were excluded. For 63% of our sample, self-reported ancestry for all grandparents was available. Ancestry was predominantly European with 96% having full European ancestry.
Both alcohol phenotypes and genotypes suitable for linkage analysis were available for 4,172 individuals (1,654 males, 2,518 females) from 1,690 families. The sample comprised 3,085 twins (including 1,465 complete pairs, of which 34 were MZ) and 1,087 nontwin siblings. The number of quasi-independent sibling pairs (QISPs – i.e., twin pairs plus pairings formed between twins and their nontwin siblings) in the dataset was 3,837. Genotyping was available for 1,338 parents (including 483 parental pairs) and these were used to refine estimates of IBD probabilities.
Participants were aged 21 to 81 years (39.7 ± 10.5) at the time of first interview. Further, 803 participants were interviewed on a second occasion, with a mean time between interviews of 10.4 ± 1.8 years (range = 2 to 13 years). Linkage analyses were run on the full sample, first, using all first interview data (Younger dataset) and second, with data from the second interview replacing first interview data for the 803 individuals interviewed twice (Older dataset).
A detailed description of zygosity determination and genotyping for this sample can be found in Hansell and colleagues (2009). For the 4,172 participants with dependence symptom data, the number of unique autosomal markers per individual ranged from 301 to 1,359 (M = 559.9 ± 234.1).
Alcohol Dependence Symptom Score
Assessment was based on lifetime prevalence. An alcohol dependence symptom score was obtained from nine items based on the substance dependence criteria from the third revised and fourth editions of the Diagnostic and Statistical Manual of Mental Disorders (American Psychiatric Association, 1987, 2000), as listed in Hansell and colleagues (2008). All items were scored Yes/No (1/0), with the exception of Items 2 and 7, which were coded as 3 level ordinal measures [Item 2: no = 0, 1 unsuccessful attempt to cut down = 1, more than 1 unsuccessful attempt to cut down = 3, and Item 7: no = 0, a marked (but less than 50%) increase in the amount required to achieve the desired effect = 1, at least a 50% increase in the amount required = 2]. Items were summed to obtain a single score. Distributions of alcohol dependence symptom scores are shown in Fig. 1. Approximately, 34% of the sample had positive responses for 3 or more criteria, consistent with being classified as alcohol dependent.
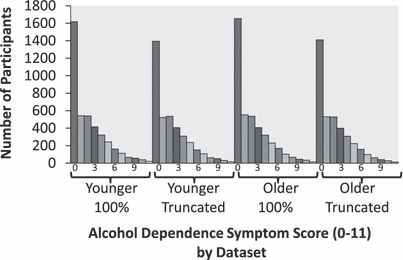
Distributions of alcohol dependence symptom scores (derived from 9 symptoms with a maximum score of 11) are shown for full (100%) and truncated samples and for datasets that differ for a subset of 803 individuals for whom data were collected on 2 occasions [first occasion data (M = 1.8 ± 2.2) were included in the Younger dataset and second occasion data (M = 2.2 ± 2.6) in the Older dataset].
Statistical Analyses
Pedigree-wide analyses were conducted for the 22 autosomal chromosomes using MERLIN-REGRESS and the X chromosome using MINX (Abecasis et al., 2002). Multipoint linkage analyses were examined for the Younger and Older datasets. Data for both co-twins from MZ pairs were modeled in analyses (with zygosity identified), although the MZ relationship per se does not contribute to linkage. Posthoc linkage analyses examined the effects of excluding individuals with the least alcohol exposure by dropping approximately 10% of the sample (8.3% for the Younger dataset and 7.6% for the Older) from the lower end of the sex and age-adjusted distribution of consumption. Essentially, this subset were nondrinkers during the previous 12-month period (<1 drink per week), although many reported having dependence symptoms at some time during their lives (reported symptoms ranged 0 to 11, M = 1.4 ± 2.8 and 1.3 ± 2.7 for the Younger and Older datasets, respectively).
Preliminary analyses were performed using SPSS (version 15.0 for Windows, 2006). The effects of sex, age, age2, sex*age, and sex*age2 were examined using stepwise linear regression. The effects of significant covariates (p < 0.05) were regressed out before linkage analyses. For the younger dataset, sex, age, age2 and their interactions (sex*age and sex*age2) were significant in both the full and truncated samples (all p values <0.001). In the older dataset, sex, age2 and sex*age were significant in the full sample (p = 0.001, 0.001, 0.009 respectively) while only sex*age (p < 0.001) was significant in the older dataset with the truncation. Note that age and year of birth correlated >0.90, and consequently, age was a good surrogate covariate to account for the effects of possible secular trends.
Normalized residuals (mean 0, variance 1) were used for all variables to facilitate analyses in MERLIN-REGRESS (Sham et al., 2002). There were no univariate outliers. Bivariate outliers, as defined in Benyamin and colleagues (2008), accounted for less than 0.4% of the sample and were excluded from linkage analyses.
Results
Multipoint linkage results are shown in Fig. 2. LOD scores exceeding 1.5 were noted on chromosome 3 (D3S1292, 140.7 cM), chromosome 4 (GATA70E01, 37.1 cM), and chromosome 5 (ATAG078, 35.1 cM; GATA134B03, 42.6 cM; and GATA7C06, 53.8 cM).
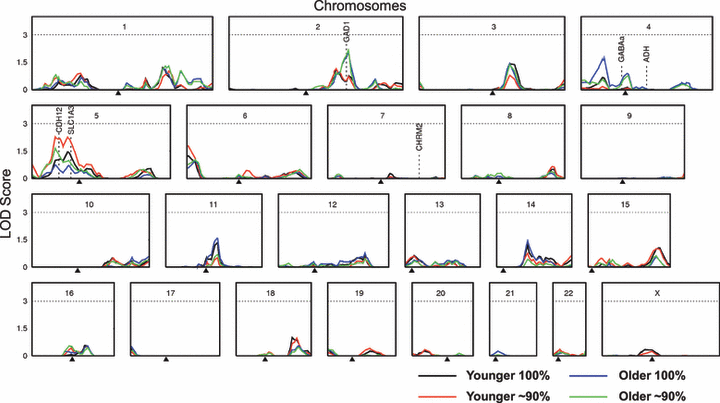
Multipoint LOD scores for alcohol dependence symptom score are shown for Younger and Older samples (identical except for a subset of 822 individuals for whom data were collected on 2 occasions with first occasion data included in the Younger dataset and second occasion data in the Older dataset) and for full (100%) and truncated (∼90%) samples. Gene locations are shown for the GABAA and ADH gene clusters and CHRM2, all of which have been associated with alcohol phenotypes in multiple studies (Edenberg and Foroud, 2006) and GAD1, SLC1A3, and CDH12, which are candidate addiction genes found under current linkage peaks. Results with map positions and marker names are shown in Table S1.
The highest multipoint LOD of 2.7 (p = 0.0002) was found on chromosome 5 using the dataset maximizing data collected at a younger age, and with lightest drinkers dropped. A similar LOD (2.6) was found when maximizing data collected at an older age. However, including light drinkers reduced maximum LOD scores in the region to 1.2 and 1.0 respectively for the younger and older samples. While none of these findings are significant by standard criteria for linkage studies, those LOD scores ≥2.2 could be considered suggestive based on an expectation of statistical evidence occurring one time at random in a genome scan (a pointwise significance level of 7 × 10−4 for sib-pair studies), as proposed by Lander and Kruglyak (1995).
Discussion
We sought to identify linkage regions for an alcohol dependence symptom score in a large community sample of Australian adults. Although none of our findings achieve genomewide levels of significance, our LOD scores greater than 2.0 are in a region on chromosome 5 previously linked to alcohol-related traits (e.g. Schuckit et al., 2001). The current findings suggest that genes in this region may influence alcohol-related problems ranging from mild through to the severe problems typically found in a clinical sample. Further, our results indicate, as previously found for alcohol consumption (Hansell et al., 2009), that the linkage method is sensitive to relatively small differences in inclusion criteria. In this case, variations in participant age and drinking status led to peak variation.
Given expected error in location estimates (Atwood and Heard-Costa, 2003), our suggestive findings converge with numerous previous reports. Linkage peaks on chromosome 5, which are maximal for the younger sample (LOD 2.7) and enhanced through exclusion of the lowest alcohol users, are approximately 10 to 15 cM from that reported for response to alcohol (Schuckit et al., 2001) and age of onset/harm avoidance/novelty seeking (Dick et al., 2002) and approximately 35 cM from a region implicated in alcoholism (Hill et al., 2004).
Genes of interest in this region include SLC1A3 and CDH12. SLC1A3 [solute carrier family 1 (glial high affinity glutamate transporter), member 3; gene map locus 5p13] is a glutamatergic neurotransmission gene with expression levels found to be significantly elevated in postmortem prefrontal cortex of alcoholic individuals compared with nonalcoholic controls (Flatscher-Bader and Wilce, 2006; Flatscher-Bader et al., 2005). CDH12 (cadherin 12, gene map locus 5p14-p13) shows increased expression in postmortem temporal cortex of individuals with a history of alcohol abuse or dependence compared with controls (Sokolov et al., 2003) and similarly, in the orbitofrontal cortex in violent suicide victims compared with nonpsychiatric controls (Thalmeier et al., 2008). Cell adhesion-related genes such as CDH12 have roles in building and maintaining synaptic structures (Benson et al., 2000), pointing to possible disturbances of synaptic neuroplasticity in individuals with alcohol dependence (Sokolov et al., 2003).
Minor linkage on chromosome 2 (LOD 1.5) is approximately 15 to 30 cM from that reported for alcoholism (Hill et al., 2004), rank-transformed alcohol dependence symptom count in the IASPSAD sample (Kendler et al., 2006), heavy alcohol use in the Heart Study (Wyszynski et al., 2003), severity of alcohol use in Mission Indians (Ehlers et al., 2004), and maximum drinks in COGA (Saccone et al., 2005). Further, it is about 40 to 50 cM from regions implicated in alcohol dependence (Reich, 1996), conduct disorder (Dick et al., 2004), and more recently, an alcohol dependence symptom score (Agrawal et al., 2008), in the COGA sample. In the current analyses, linkage was maximal in the older sample and sample truncation did not alter the result.
This region of chromosome 2 is home to GAD1 (glutamate decarboxylase 1, gene map locus 2q31), which catalyzes the conversion of glutamic acid to gamma-aminobutyric acid (GABA), the major inhibitory neurotransmitter in the vertebral central nervous system. It has been associated with alcohol dependence in Han Taiwanese males (Loh et al., 2006). In the IASPSAD sample, no association was found with dependence, but significant associations were found with initial sensitivity to alcohol and to age at onset of dependence (Kuo et al., 2008). Kuo and colleagues (2008) proposed that the underlying pathophysiology regulated by GAD1 may be more directly related to the component processes of dependence, than to the disorder itself.
A small linkage peak (LOD 1.6) on chromosome 3 was located in the same region as linkage found previously in the same sample for alcohol consumption (LOD 2.7) (Hansell et al., 2009). In addition, a minor peak (LOD 1.0) was observed in the region encompassing the gamma-aminobutyric acid receptor A subunit 2 (GABRA2) gene. This gene and others in its cluster on chromosome 4 (GABRA4, GABRB1, GABRAG1) have been found to be associated with alcohol dependence (as well as other substance use disorders and psychopathology) across several independent studies (Covault et al., 2004; Edenberg et al., 2004; Fehr et al., 2006; Lappalainen et al., 2005; Lind et al., 2008; Matthews et al., 2007; Soyka et al., 2008).
Some limitations need to be considered. First, these results were not genomewide significant. However, the likelihood that these are false positives is somewhat diminished as the results replicate linkage found in these regions by previous studies. Further, quantitative trait loci of small effect are to be expected given that an alcohol dependence symptom score is likely to be influenced by many genes of small effect, as has been found for numerous traits in genomewide association studies (Wray et al., 2007). Larger linkage signals, as have been reported previously, may simply reflect the high variance of effect size estimates found with smaller samples. Linkage differences related to sample variation were found in the current study, but are not sufficiently large for meaningful interpretation. A further limitation is that the component samples were collected for heterogeneous purposes and may not be entirely representative of the adult Australian population. Notwithstanding these limitations, our analyses demonstrate the utility of a community sample, results of which are easily generalizable, for linkage analyses. The chromosomal regions identified provide a focus for future gene-mapping efforts.
Acknowledgments
We thank all participating families, the project coordinators and interviewers under the supervision of Dixie Statham, the data managers under the supervision of John Pearson and David Smyth, and laboratory personnel under the supervision of Megan Campbell and Anjali Henders. For genome scans of twins and siblings, we acknowledge and thank the Mammalian Genotyping Service, Marshfield WI (Director: Dr. James Weber) for genotyping under a grant to Dr. Daniel O’Connor; Drs. Eline Slagboom and Dorret Boomsma for the Leiden genome scan; Dr. Peter Reed for the Gemini genome scan; and Dr. Jeff Hall for the Sequana genome scan. National Institute of Health grants have supported this work, including grants to Drs. Andrew Heath (AA07728, AA10248, AA11998, AA13321), Pamela Madden (DA12854), Nick Martin (AA13326), Michele Pergardia (DA019951), Richard Todd (AA13320), and John Whitfield (AA014041). Dr. Arpana Agrawal receives support from DA023668 and ABMRF/The Foundation for Alcohol Research and Dr. Katherine Morley from a National Health and Medical Research Council Public Health Fellowship (520452). With the exception of ABMRF, all funding is government sourced. Funding bodies played no role in the collection or analysis of data nor in the writing of this paper.




