Diverged and conserved aspects of heart formation in a spider
Abstract
SUMMARY Heart development exhibits some striking similarities between vertebrates and arthropods, for example in both cases the heart develops as a linear tube from mesodermal cells. Furthermore, the underlying molecular pathways exhibit a significant number of similarities between vertebrates and the fruit fly Drosophila, suggesting a common origin of heart development in the last common ancestor of flies and vertebrates. However, there is hardly any molecular data from other animals. Here we show that many of the key genes are also active in heart development in the spider Cupiennius salei. Spiders belong to the chelicerates and are distantly related to insects with respect to the other arthropods. The tinman/Nkx2.5 ortholog is the first gene to be specifically expressed in the presumptive spider heart, like in flies and vertebrates. We also show that tinman is expressed in a similar way in the beetle Tribolium castaneum. Taken together this demonstrates that tinman has a conserved role in the specification of the arthropod heart. In addition, we analyzed the expression of other heart genes (decapentaplegic, Wnt5, H15, even-skipped, and Mef2 ) in Cupiennius. The expression of these genes suggests that the genetic pathway of heart development may be largely conserved among arthropods. However, a major difference is seen in the earlier expression of the even-skipped gene in the developing spider heart compared with Drosophila, implying that the role of even-skipped in heart formation might have changed during arthropod evolution. The most striking finding, however, is that in addition to the dorsal tissue of the fourth walking leg segment and the opisthosomal segments, we discovered tinman-expressing cells that arise from a position dorsal to the cephalic lobe and that contribute to the anterior dorsal vessel. In contrast to the posterior heart tissue, these cells do not express the other heart genes. The spider heart thus is composed of two distinct populations of cells.
INTRODUCTION
Complex multicellular organisms require a system for the distribution of nutrients, transport of gas, and transport of excretions. The circulatory system therefore is a key invention during evolution. The insect and vertebrate hearts are assumed to be homologous structures based on the similarity of the underlying molecular pathways that regulate the development of the tissue in both the fruit fly Drosophila and vertebrates (Bodmer and Venkatesh 1998; Frasch 1999; Chen and Fishman 2000). The vertebrate blood circulatory system including the heart and the arthropod dorsal vessel (heart) are both mesodermal structures. A further similarity between both groups is that the heart initially forms as a linear tube.
In arthropods the heart forms from the dorsal mesoderm. In the fly Drosophila melanogaster, the transcription factors Twist (Twi) and Snail (Sna) specify the mesoderm. Twist activates mesodermal genes whereas Snail represses non-mesodermal genes (Leptin 1991). Work in the beetle Tribolium castaneum and the spider Achaearanea tepidariorum shows that the function of twi in mesoderm determination and specification seems to be conserved between insects and spiders (Sommer and Tautz 1994; Handel et al. 2005; Yamazaki et al. 2005). In contrast to that, the function of sna in mesoderm formation seems to be less conserved among insects and spiders (Yamazaki et al. 2005). After gastrulation, the mesoderm of Drosophila forms a cell layer that extends from the ventral midline to the dorsal edge of the germ band (Klapper et al. 1998). When the dorsal movement of the mesoderm has come to an end, clusters of heart progenitors arise in the most dorsal region of the mesoderm on both sides of the Drosophila germ band and form a proper tube, the dorsal vessel (the heart) (Rizki 1978; Cripps and Olson 2002; Zaffran and Frasch 2002).
An important factor in Drosophila heart development is the homeobox containing transcription factor Tinman (Tin), which is a direct target of Twi (Bodmer et al. 1990; Yin et al. 1997). Loss of tin function results in a loss of dorsal mesodermal structures including cardioblasts and pericardial cells (Azpiazu and Frasch 1993; Bodmer 1993). Expression of tin starts just before gastrulation and it is initially observed in all cells of the prospective mesoderm. After gastrulation, tin expression is restricted to the dorsal mesoderm, where it makes this tissue competent to form visceral mesoderm, dorsal muscles, and heart cells (Azpiazu and Frasch 1993; Bodmer 1993; Yin et al. 1997). Although tin plays an important role in this process, its expression is not sufficient to form dorsal mesodermal structures. At least one other gene, pannier (pnr) is required to activate heart-specific genes, together with tin (Gajewski et al. 1999). The expression of tin in the dorsal mesoderm is transient and becomes restricted to the heart progenitors where it continues to be expressed as the heart matures.
Decapentaplegic (dpp) and wingless (wg) are responsible for maintenance of tin expression in dorsal structures of the mesoderm in Drosophila. Both genes encode secreted signalling molecules. dpp is expressed in the ectodermal tissue and is thought to give the mesoderm a segmented pattern (Staehling-Hampton et al. 1994). No dorsal vessel forms in the absence of Dpp activity in the ectodermal tissue overlaying the dorsal mesoderm (Seidel et al. 1940; Frasch 1995). This indicates that Dpp is an essential inductive signal for heart development in the fly. However, ectopic expression of tin and dpp is not sufficient to control heart formation (Rugendorff et al. 1994; Frasch 1995). wg is expressed in segmentally repeated patterns in the ectoderm overlaying the tin expressing domains. Subsequently the heart progenitors are established beneath each wg stripe (Jagla et al. 1997). After this step these cells differentiate into cardiac cells, pericardial cells, and body wall muscle progenitors.
In Drosophila two T-Box genes, midline (mid) and H15, which are related to the vertebrate Tbx20 genes, are also known to play an important role in heart development (Miskolczi-McCallum et al. 2005; Reim et al. 2005). The H15/mid T-Box genes are required for the diversification of cardiac cells (Qian et al. 2005; Miskolczi-McCallum et al. 2005; Reim et al. 2005).
Another gene that acts in the differentiation of cardiac cells in Drosophila is the MADS-box gene myocyte enhancing factor 2 (DMef2) (Gajewski et al. 1997; Frasch 1999). It is a direct target of tin and is expressed in all cardiac, somatic, and visceral muscle cell precursors during mesoderm formation. DMef2 function is required for the differentiation of heart, body wall, and gut muscle cells. In the heart, DMef2 is expressed in all contractile cardiac cells where it activates the transcription of the myosin subunit genes. Cardiac cells form in embryos mutant for DMef2 but do not properly differentiate (Bour et al. 1995; Lilly et al. 1995; Ranganayakulu et al. 1995).
The vertebrate heart is also formed from bilaterally symmetrical precursors of mesodermal origin and Nkx2.5, BMP2/4, and GATA genes seem to play similar roles in heart development as their Drosophila orthologs tin, dpp, and pnr respectively (reviewed in Bodmer and Venkatesh 1998; Chen and Fishman 2000). T-Box genes and Mef2 genes are also involved in heart formation in vertebrates (Doevendans and van Bilsen 1996; Griffin et al. 2000; Meins et al. 2000; Miskolczi-McCallum et al. 2005; Plageman and Yutzey 2005; Reim et al. 2005). Therefore, the genetic network specifying the heart field in vertebrates and the heart tissue in Drosophila appears to include many conserved characteristics that may date back to their last common ancestor (e.g., Zaffran and Frasch 2002). However, very little information is available about the genes involved in heart formation in other animals. A better understanding of the ancestral mechanisms for heart development could contribute to better understand human heart diseases caused by mutations in genes in this conserved genetic cascade, such as for instance the Holt-Oram and DiGeorge syndromes that are caused by mutations in T-Box genes (reviewed in Plageman and Yutzey 2005).
To obtain a better understanding of the evolution of heart development within arthropods and between these animals and other metazoans, such as vertebrates, we analyzed orthologs of key genes involved in Drosophila heart development in the spider Cupiennius salei. Orthologs of tin/Nkx2.5, dpp/BMP2/4, wg/Wnt, eve/evx, H15/TBX20, and DMef2/Mef are all expressed in the developing dorsal vessel of this spider. The temporal dynamics of the expression of these genes point to many similarities but also to some differences in heart development between insects and spiders.
MATERIAL AND METHODS
Cloning of spider genes
Gene fragments were recovered via PCR-based screens on cDNA prepared from total RNA of either C. salei or T. castaneum embryos (Damen et al. 2000; Prpic et al. 2003; Damen et al. 2005). cDNA was prepared in a reverse transcription reaction using the Superscript First Strand Synthesis System for RT-PCR (Invitrogen, Karlsruhe, Germany). The degenerate primers used in the PCR reactions are based on conserved amino acid sequences, and were purchased from Metabion (Martinsried, Germany). For tin we used the primers tin-fw (CAN CCN TTY WSN GTN AAR GA) and tin-bw-1 (RAA CCA DAT YTT NAC YTG NGT) in a first PCR, and tin-fw and tin-bw-2 (TG NGC YTG NSW RAA NAR NAC) in a nested PCR. For Mef2 we used Mef2-fw (ATG GGN MGN AAR AAR ATH CA) and Mef2-bw (TCR TGN GGY TCR TTR TAY TC). Additional sequence information of Cs-Mef2 was obtained via 3′-RACE-PCR. A fragment of the H15 gene of Tribolium was isolated via PCR using the primers as described in Prpic et al. (2003). The fragments have been sequenced on both strands; the sequences are available under accession numbers AM749796 (Cs-Mef2), AM749797 (Cs-tin), AM749798 (Tc-tin), and AM888353 (Tc-H15). Other sequences have been published previously (Nagy and Carroll 1994; Sanchez-Salazar et al. 1996; Damen et al. 2000; Damen 2002; Prpic et al. 2003) and are available under accession numbers AJ252155 (Cs-eve), AJ315946 (Cs-Wnt5-1), AJ518936 (Cs-dpp), AJ518938 (Cs-H15-1), AJ518939 (Cs-H15-2), U63132 (Tc-dpp), and AAB29938 (Tc-wg).
Fixation and preparation of embryos
The Cupiennius embryos were obtained from our lab culture in Cologne (Damen et al. 2005) and were dechorionated in 50–100% bleach for 1–2 min and fixed with 5% formaldehyde in heptane for 16 h. After exchanging the fixative with methanol, the vitelline membrane was manually removed using Dumont-5 forceps (Damen and Tautz 1998).
A series of developmental stages was prepared by fixation of embryos of one single cocoon at 6 h intervals. Usually all embryos within a cocoon develop simultaneously.
In situ hybridization
Whole-mount in situ hybridization with DIG labelled RNA probes was carried out as described for Drosophila (Tautz and Pfeifle 1989) and modified after Damen and Tautz (1998) for Cupiennius and after Wolff et al. (1995) for Tribolium.
RESULTS
Isolation of genes involved in heart formation
Orthologs of genes known to be involved in heart formation in vertebrates and the insect Drosophila were recovered from the spider C. salei and the beetle T. castaneum by RT-PCR.
We isolated a 556-bp fragment of the tin gene from the spider and a 325-bp fragment from the beetle using primers directed against sequences in the TIN domain and the homeodomain. The sequence of the recovered fragments shows strong similarity to the Drosophila tin and the vertebrate Nkx2.5 genes and therefore are designated as Cs-tinman (Cs-tin), and Tc-tinman (Tc-tin), respectively (Fig. 1A).
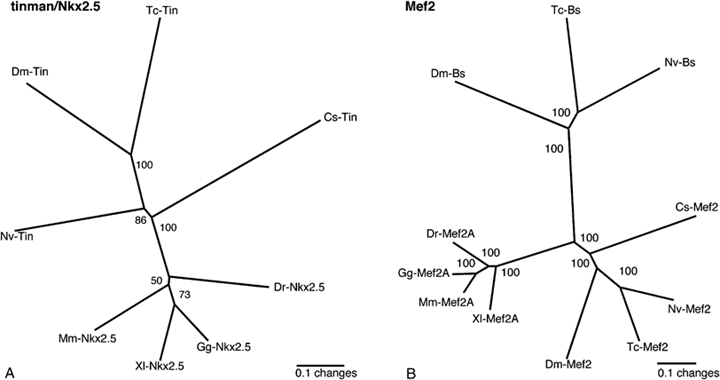
Phylogenetic analysis of Tinman and Mef2 gene fragments. Shown is the unrooted Puzzle tree computed from 1000 intermediate trees produced with the Quartet Puzzling method (Strimmer and von Haeseler 1996). The numbers at the tree edges are the reliability values. (A) Arthropod and vertebrate Tinman/Nkx2.5 sequences and (B) arthropod and vertebrate MEF2 and Blistered (Bs) sequences. Cs, Cupiennius salei; Tc, Tribolium castaneum; Dm, Drosophila melanogaster; Nv, Nasonia vitripennis; Mm, Mus musculus; Xl, Xenopus laevis; Gg, Gallus gallus; Dr, Danio rerio.
We also isolated a spider ortholog of the Drosophila DMef2 and vertebrate Mef2 genes (Fig. 1B). The recovered fragment is 1952-bp long and codes nearly the complete ORF including the major part of the characteristic N-terminal MADS domain and the complete MEF domain. The fragment contains a 3′UTR including a poly-A stretch. Based on sequence comparisons with Drosophila and vertebrate sequences, we assume that seven amino acids are missing at the N terminus of the Cs-Mef2 sequence.
From Tribolium we also recovered a fragment of a T-Box gene with high similarity to the Drosophila gene H15 that we designated as Tc-H15.
Expression of tinman in the heart precursors of the spider and the beetle
In Drosophila, tin is the first gene expressed in the mesodermal tissue that will give rise to the heart (Bodmer 1993). It is activated by Twist, which activates the transcription of mesoderm-specific genes (Yin et al. 1997). To study the formation of the heart in the spider we first analyzed the expression of the tin gene.
The spider body consists of two tagmata: an anterior prosoma, which includes the six appendage-bearing segments, and a posterior opisthosoma. The heart lies dorsal in the anterior of the opisthosoma and is formed from cells at the dorso-lateral edge of the germ band stage embryo (Wilson 1967; Anderson 1973; Foelix 1996). The description of the expression patterns below focuses on the expression in the presumptive heart region. Other aspects of the expression and the function of these genes in segmentation and leg development have been described previously (Damen et al. 2000, 2005; Damen 2002; Prpic et al. 2003).
Cs-tin transcripts are detected at the dorsal-most edge of the opisthosomal segments in the germ band embryo (Fig. 2, A and B). Cs-tin is initially expressed at the dorso-lateral edge of each segment (Fig. 3D) but later is in a continuous stripe at the dorso-lateral edge of the embryo (2, 3). The anterior border of Cs-tin expression, however, extends into the segment that bears the fourth walking leg (L4) where it has a sharp expression boundary in the posterior portion of this segment (2, 3). Using the position of the leg on this segment as a landmark, it is clear that this boundary corresponds to the parasegment boundary (Damen 2002). The terminal growth zone at the posterior end of the embryo is free of tin transcripts (Fig. 2A). Cs-tin is expressed in the dorso-lateral edge of these segments, until completion of dorsal closure when the heart tube has formed (Fig. 3, A–C). Surprisingly, during inversion that eventually leads to dorsal closure, additional Cs-tin expression is seen in cells at a position dorsal to the cephalic lobe (Fig. 3A). The origin of these tin-expressing cells remains unclear; presumably they originate from the ridge of the dorsal cephalic lobe, but we cannot exclude that these cells are recruited from extra-embryonic tissue located just dorsally to the cephalic lobe. During completion of the inversion and dorsal closure, it becomes obvious that these cells contribute to the anterior part of the developing heart (Fig. 3, B and C).
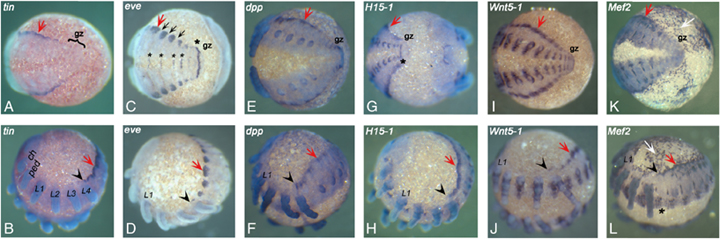
Expression of tin, eve, dpp, H15, Wnt5-1, and Mef2 orthologs in the developing heart of the spider Cupiennius salei. Expression of tin (A, B), eve (C, D), dpp (E, F), H15-1 (G, H), Wnt5-1(I, J), and Mef2 (K, L). The red arrows point to expression of these genes in the dorso-lateral edge of the germ band, which will form the heart. The black arrowheads mark the anterior border of the dorsal expression, which is in all cases except for Mef2 in the posterior of the fourth walking leg segment. (A) The posterior growth zone is free of tin transcripts. (C, D) Expression of even-skipped (eve) in dorsal patches (red arrow) along the dorso-lateral edge separated by tissue that does not express eve (black arrows). Asterisks mark expression in the neural ectoderm and in the growth zone. (G) Asterisk marks stripe of Cs-H15-1 expression in the last newly formed segment. (K, L) Myocyte enhancing factor 2 (Mef2) expression at the dorso-lateral edge extends less far posterior than for the other genes at this stage. Expression in the extra-embryonic tissue (white arrow). Furthermore, the anterior border is in the first opisthosomal segment (black arrowhead) and not in L4 as for the other genes. The asterisk marks Cs-Mef2 expression in the developing ventral nervous system. Note that uniform expression of Cs-dpp and Cs-Mef2 in the appendages is artifactual and due to elongated staining. A, C, E, G, I, K are ventral views; B, D, F, H, J, L are lateral views. In all cases anterior is to the left. ch, chelicere; gz, growth zone; L1–L4, walking legs 1–4; ped, pedipalp.
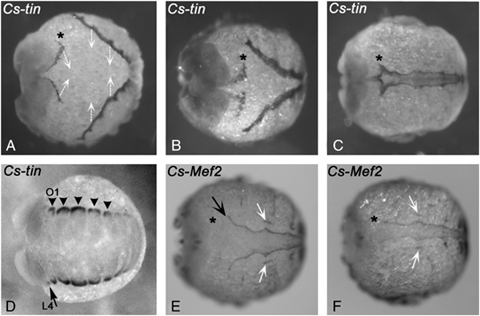
Expression of spider tinman and Mef2 during inversion and dorsal closure. Dorsal views of late embryonic stage showing dorsal closure. (A–C) Cs-tin expression; (E, F) Cs-Mef2 expression. The dark staining represents expression of Cs-tin and Cs-Mef2 in the dorsal edge of the germ band that moves over the yolk to close at the dorsal side. (A) The white arrows denote the direction of germ band movement during inversion. The asterisk marks anterior Cs-tin expressing tissue dorsal to the head lobe. (B) Cs-tin expressing tissue converges to the dorsal midline of the embryo. Asterisk as in A. (C) Dorsal closure. tin expressing cells from both sides of the split germ band meet and fuse along the dorsal midline. The proper heart tube forms. Asterisk as in A. (E, F) Expression of Cs-Mef2 in embryo short before (E) and at (F) dorsal closure. Cs-Mef2 is expressed in the developing heart (black arrows) and in the alary muscles besides the heart (white arrows). Note that Cs-Mef2 is not expressed in the Cs-tin positive cells dorsal to the head lobe (asterisks). (D) Early expression of Cs-tin in dorsal mesodermal blocks that will give rise to the later heart tube. Arrowheads point to the blocks of expression. Arrow marks expression in the posterior part of the L4 segment. L4, fourth walking leg segment; O1, first opisthosomal segment.
Expression of Cs-eve, Cs-dpp, Cs-H15-1, Cs-H15-2, Cs-Wnt5-1, and Cs-Mef2 in the spider heart precursors
The expression of Cs-tin suggests that it specifies the heart in the spider, as it does in vertebrates and insects. To further explore the gene network involved in heart formation we analyzed the expression of spider orthologs of additional genes, which are known to play a role in Drosophila heart formation.
The spider even-skipped gene (Cs-eve) is expressed along the dorso-lateral edge of the germ band stage embryo in a very similar pattern to Cs-tin (Fig. 2, C and D). The segmental expression as seen in Fig 2, C and D is due to a slightly earlier developmental stage of the embryo shown here and does not resemble an exceptional characteristic for Cs-eve. Also other early heart genes are initially segmentally expressed (not shown). Transcripts of Cs-eve become first detectable in the spider heart about 6–12 h after initial Cs-tin expression (Fig. 5) and persist until dorsal closure has been completed (not shown). The dorsal Cs-eve expression has the same anterior expression boundary in L4 as Cs-tin (Fig. 2, B and D). Neither Cs-eve nor any of the other analyzed genes (dpp, H15, Wnt5-1, Mef2) can be detected in the Cs-tin positive cells that are dorsal to the cephalic lobe (e.g., 2, 3).
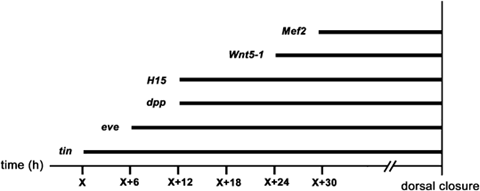
Overview of the temporal appearance of gene expression in the developing heart of Cupiennius salei. Embryos of a single cocoon were fixed at 6-h intervals. The first gene to be expressed in the presumptive heart is Cs-tin. This time point is defined as time point “X” and corresponds to about 8 days of embryonic development, the other time points are relative to time point X (in hours).
Cs-dpp is expressed at the dorso-lateral edge where the heart precursors form at about 12 h after the appearance of Cs-tin (2, 5). The anterior-most Cs-dpp expression is in the posterior part of the L4 segment and extends throughout the opisthosomal segments to the last formed segment anterior to the growth zone (Fig. 2E). Expression in the more posterior segments is weaker than in the anterior ones. Cs-dpp expression persists until the heart vessel has formed after dorsal closure (not shown). Note that detection of dpp transcripts in the heart required prolonged staining that resulted in overstaining of the appendages (compare with Prpic et al. 2003).
The Cs-H15-1 gene, a Tbx-20 like T-box gene, is expressed at the same time as Cs-dpp at the dorso-lateral edge in the cells forming the spider heart (2, 5). In addition, the Cs-H15-1 expression extends from the posterior half of the L4 segment (Fig. 2, G and H) to the nascent segment anterior to the growth zone (Fig. 2G) and is maintained until the heart tube has formed after dorsal closure (not shown). Cupiennius actually contains two paralogs of the Tbx-20-like T-Box gene H15 (Prpic et al. 2003). The second paralog, Cs-H15-2, shows the same expression pattern in the presumptive heart as Cs-H15-1 (not shown).
About 12 h after the initial onset of Cs-dpp and Cs-H15 expression in the spider heart, Cs-Wnt5-1 is activated (2, 5). Cs-Wnt5-1 expression is also observed in the dorsal region of the Cupiennius embryo from the posterior part of the fourth leg-bearing segment L4 (Fig. 2J) to the nascent opisthosomal segment anterior to the growth zone (Fig. 2I). Cs-Wnt5-1 expression continues until dorsal closure is completed (not shown).
Cs-Mef2 expression appears about 6 h after the first appearance of Cs-Wnt5-1 in the heart (2, 5). As Mef2 genes are known to be responsible for muscle cell differentiation we assume that the complex expression profile of Cs-Mef2 reflects activity in muscle precursors of the somatic, visceral and cardiac mesoderm and is associated with the differentiation of contractile cells. Cs-Mef2 expression is clearly visible in the heart tube of Cupiennius up to dorsal closure (2, 3). The anterior most dorsal expression domain of Cs-Mef2, however, is located in the first opisthosomal segment—one segment more posterior than the expression of the other genes described here. In addition, it does not extend as far posterior as the other genes at this stage (Fig. 2K). We also detected Cs-Mef2 expression in the developing ventral nervous system in a pattern similar to the pattern of pro-neural genes (Fig. 2L) (Stollewerk et al. 2001) suggesting Cs-Mef2 has a role in CNS development. Drosophila DMef2 is known to play a role in neural cell differentiation (Schulz et al. 1996), as is Mef2 of the sea anemone Nematostella (Martindale et al. 2004) and vertebrates (e.g., Skerjanc and Wilton 2000). Thus this data suggests an ancestral function of Mef2 genes in arthropod neural cell development. Cs-Mef2 is also expressed in the extra-embryonic tissue but not in the tissue between the splitting ventral germ band (Fig. 2, K and L).
Expression of Tc-tin, Tc-dpp, Tc-H15, and Tc-wg in the heart precursors of the beetle T. castaneum
We also analyzed the expression in the heart precursors of a number of genes involved in Drosophila and Cupiennius heart development in the short germ beetle T. castaneum. This beetle develops segments sequentially from the posterior growth zone, like in the spider and most other arthropods, but unlike Drosophila where all segments form simultaneously at the blastoderm stage (e.g., Davis and Patel 2002).
Like the spider tin ortholog, the Tribolium tin gene is exclusively expressed in the heart precursors (Fig. 4, A and B). Expression is seen in the mesoderm at the dorsal edge of the germ band stage embryo. Tc-tin expression persists until dorsal closure like Cs-tin (not shown). The anterior most border of Tc-tin expression is in the labial segment (Fig. 4A). The posterior expression extends up to the most nascent opisthosomal segment. There is also Tc-tin expression in the cephalic lobe, in a position comparable with the reported anterior expression in Cupiennius. (Fig. 4, A and B).

Expression of tinman, decapentaplegic, H15 and wingless in the beetle Tribolium castaneum. (A) Flat-mounted germ band stage embryo of Tribolium castaneum with ten abdominal segments. Expression of Tc-tin is in the mesoderm at the dorso-lateral edge. Tc-tin expression expands from the labial segment (arrow) to the last formed segment. At this stage expression is still restricted to mesoderm blocks along the dorsal edges of the embryo. Note that there is additional expression of Tc-tin at the dorsal edge of the cephalic lobe (arrowheads). (B–E) Flat-mounted germ band retraction stage embryos of Tribolium stained for Tc-tin (B), Tc-dpp (C), Tc-H15 (D), and Tc-wg (E). Arrowheads in (B) point to cephalic expression of Tc-tin; asterisks in (E) demarcate patches of wg expression in the dorsal ectoderm. A1 and A10, abdominal segments 1 and 10; ect, ectoderm; lb, labial segment; lr, labrum; mes: mesoderm; T1, first thoracic segment; tel, telson.
The beetle T-box gene Tc-H15 is expressed in the dorsal mesoderm at a comparable position in the heart precursors to its ortholog in the spider (Fig. 4D). The Tribolium dpp gene is expressed in ectodermal tissue at the dorsal edge of the embryo overlying the Tc-tin and Tc-H15 expressing cells (Fig. 4C), Tribolium wg expression is also present in segmental spots at the dorsal edge (Fig. 4E).
Shared anterior border of expression and temporal order of expression during heart development in the spider
The anterior border of expression of the genes used in this study is in the posterior half of the segment that bears the fourth walking leg (L4) (Fig. 2). Using the leg as a landmark, it is clear that the position of this expression border corresponds to the parasegment boundary (Damen 2002). The only exception is Cs-Mef2, which has its anterior expression border one segment more posterior. The anterior expression boundary of these genes does not change during development; we never found any variation in the sharp anterior expression border for all the examined genes in the heart (Fig. 2).
The temporal aspects of expression are summarized in Fig. 5. Starting at the germ band stage, embryos of a single cocoon have been fixed in intervals of 6 h and used for in situ hybridization for the different genes. The first gene to be expressed in cells at the dorso-lateral edge that will form the heart is Cs-tin (defined as time point X in Fig. 5; X corresponds to about 8 days after egg laying). About 6 h later (X+6), expression of Cs-eve becomes visible at the dorso-lateral edge, and again 6 h later (X+12) expression of Cs-dpp and Cs-H15-1/2 is first visible. Cs-Wnt5-1 expression is detectable another 12 h later (X+24), while the initial detection of Cs-Mef2 expression in the heart precursors is at around 30 h after initial expression of tin (X+30) (Fig. 5).
DISCUSSION
In both vertebrates and arthropods, the heart structures congregate into a linear tube from cells of mesodermal origin, using similar genetic pathways (Bodmer and Venkatesh 1998; Frasch 1999). However, apart from chordates and the fruit fly there was a paucity of molecular data on heart development from other animals. Our study shows that a number of the key genes are also involved in formation of the heart vessel in a spider and point to a largely conserved genetic pathway for heart development in arthropods. Furthermore, this data supports the idea of a common origin of the vertebrate and arthropod heart. tin/Nkx2.5, dpp/BMP-2/4, H15/TBX20, and DMef2/Mef2 play a role in heart formation in both vertebrates and arthropods (Bodmer and Venkatesh 1998; Griffin et al. 2000), while eve/evx and wg/Wnt seem to be involved only in heart formation in arthropods (Bodmer and Venkatesh 1998). A remarkable difference is the contribution of a distinct population of dorsal cells from the anterior cephalic lobe to the heart of the spider.
tinman is an early marker for heart development in arthropods
The tin/Nkx2.5 ortholog Cs-tin is the first gene that can be detected specifically at the dorso-lateral edge of the spider embryo at the germ band stage. These cells meet at dorsal closure and form the tube of the dorsal vessel or heart (Anderson 1973; Foelix 1996). The tin gene of the beetle Tribolium is expressed in a similar pattern at the dorso-lateral edge of the germ band. These data may imply that both in the spider (chelicerate) and the beetle (insect) the tin gene is important for the specification of the heart, as it is in Drosophila and vertebrates (Azpiazu and Frasch 1993; Bodmer 1993; Zaffran and Frasch 2002). Although other factors like twist and DMef2 are expressed earlier in the tissue that will give rise to the heart in the fly, tin is the first gene that is specifically expressed in cardiac tissue (Azpiazu and Frasch 1993; Bodmer 1993). Our data show that tin expression is exclusively in the tissue that develops into the heart in distantly related arthropods (the chelicerate Cupiennius and the insects Tribolium and Drosophila). They suggest that tin is required for heart specification in all arthropods and thus is an excellent molecular marker to track heart development.
Two different cell populations contribute to the spider heart
One of the most remarkable findings in the spider is the contribution of cells from the dorsal edge of the anterior cephalic lobe to the heart tube. None of the other genes that contribute to the development of the heart are expressed in this anterior tissue at any time. However, during inversion and dorsal closure it becomes clear that these cells contribute to the anterior portion of the dorsal vessel (Fig. 3). How these cells become specified and what activates tin in this anterior domain remains to be solved, but the maintenance of tin expression in these cells cannot be regulated by Dpp/Wg as these genes are not expressed in these cells. Furthermore, the absence of genetic factors responsible for the differentiation of heart cells, like eve, H15/mid, and Mef2, suggests that differently specified tissue contributes to this anterior most part of the dorsal vessel. Thus at least two different tissues contribute to the dorsal vessel in the spider: (1) Cells from the dorsal edge of the cephalic lobe that only express tin and (2) cells from the dorsal edge of the opisthosoma that express a similar set of genes as the developing heart in Drosophila, including tin, eve, dpp, H15, Wnt5-1, and Mef2.
In the beetle Tribolium we also detected anterior cephalic Tc-tin expression, which shows that the contribution of cells from the dorsal cephalic lobe to the forming dorsal vessel is not a peculiarity of spiders but in fact there may be a similar contribution of anterior dorsal cells to the heart in other arthropods. The contribution of the posterior cells to the heart seems to be highly conserved among arthropods. Heart precursors become specified at identical positions from the lateral dorsal edge of the germ band embryo in the spider and in insects. They presumably are specified by a similar genetic network in both groups. The genetic network for the anterior cells, however, appears to be different.
Remarkable, while the anterior expression border of Cs-Mef2 is in the posterior of the first opisthosomal segement (O1) of the spider, the anterior expression border of the other heart expressing genes is one segment more anteriorly (posterior portion of L4). Mef2 genes activate contractile protein genes (Bour et al. 1995; Lilly et al. 1995; Ranganayakulu et al. 1995; Gajewski et al. 1997). Assuming that the function of Mef2 genes in specifying contractile cells is conserved between the fly and the spider, this means that the anterior-most part of the heart tube, the anterior aorta (Anderson 1973; Foelix 1996), may lack contractile cells. Alternatively, the anterior aorta may develop contractive cells later or in a Mef2-independent manner.
Regulatory network of heart formation
The present work shows that many of the genes that act in the tin regulatory network of heart formation in Drosophila are also expressed in the presumptive heart field of the spider (Fig. 2). Although functional studies are lacking, the sequence in which the different genes become expressed (Fig. 5) gives some clues on the genetic interactions that act in spider heart formation.
Slightly later than tin, dpp is expressed in the same region as the tin-expressing cells. DPP signalling might have a role here in maintaining the tin expression during spider heart development, as ectodermal Dpp maintains tin expression in heart precursors of Drosophila (Frasch 1995; Yin et al. 1997). Indeed Tribolium dpp is also expressed at the dorso-lateral edge (Fig. 4C) (also, e.g., van der Zee et al. 2006) indicating that the role of dpp signalling is conserved in arthropod heart development. Furthermore, the function of Dpp in maintaining tin seems to be evolutionary conserved as it is also observed in vertebrates where the dpp ortholog BMP2 is required for maintaining expression of the tin ortholog Nkx2.5 (Brand 2003).
Cs-Wnt5-1 is expressed later than dpp, which is consistent with the regulatory network in Drosophila, where the combinatorial activity of tin, dpp, and wg is required for the segmental formation of heart precursors in the dorsal mesoderm (Frasch 1995; Wu et al. 1995). The sequential expression of Cs-tin, Cs-dpp, and Cs-Wnt5-1 during spider heart development suggests a conservation of the regulatory interaction of these genes in spiders and insects. In contrast to the insects Drosophila and Tribolium and the crustacean Mysidium columbiae (e.g., Wu et al. 1995; Duman-Scheel et al. 2002), it is not the spider ortholog of Wnt-1/wg that is expressed in the dorsal vessel of the spider, but a Wnt-5 ortholog. On the other hand, Wnt5 is not expressed in the developing heart of Tribolium (Bolognesi et al. 2008). Previously it has been proposed that Cs-Wnt5-1 performs the role of wg as segment-polarity gene in the ventral area of the segments of the spider (Damen 2002). The expression in the presumptive heart suggests a second case where this spider Wnt-5 gene fulfils a role that is performed by Wnt-1/wg in insects and crustaceans. Interesting, in the myriapod Glomeris marginata, like in the spider it also the Wnt5 ortholog and not the wg ortholog that is expressed in the heart (Janssen et al. 2004).
In Drosophila the combinatorial activity of tin, dpp, and wg activates lethal of scute (l'sc), which is confined to a smaller cluster of cells that then will express eve. The first appearance of eve in the heart region of the spider, however, is clearly before dpp and Wnt expression, which points to a modified or additional role of eve in spider heart development. There is no known involvement of the vertebrate eve ortholog evx in vertebrate heart development; it is thus impossible to draw conclusions on the ancestral role of eve in metazoan heart formation at this point except that the involvement of eve at least seems to date back to the last common ancestor of the arthropods. It is also not known if there is a l'sc homolog in the spider. Cs-Ash1 and Cs-Ash2, the spider orthologs of genes of the Drosophila achaete/scute complex, are not expressed in the heart region and no other candidate gene has been found (Stollewerk et al. 2001). Thus although the tin–dpp–Wnt genetic network for heart development may be conserved, the role of eve and a possible l'sc appears to be modified. Further research is required to investigate the exact role of eve in spider heart development.
The spider H15 T-box factors are expressed in developing heart tissue, similar as their Drosophila orthologs H15 and midline and the vertebrate Tbx20 gene orthologs (Miskolczi-McCallum et al. 2005; Plageman and Yutzey 2005; Reim et al. 2005). One of the major functions of mid and H15 in Drosophila is the re-activation of tin expression during cardioblast development at a stage when the induction of tin by Dpp in the dorsal mesoderm has ceased. These T-box genes are required for the normal functional diversification of cardioblasts and the expression of tin-dependent terminal differentiation genes within the dorsal vessel (Reim et al. 2005). Although we do not know the exact role of H15 in spider cardiac development, the expression patterns points to an involvement of the T-box factors in spider heart development.
Early determination of heart tissue
The spider tin gene is the first gene that can be specifically detected at the dorso-lateral edge of the embryo. In Drosophila, tin is directly activated by twi and is required for the specification of the dorsal mesoderm, including the heart (Yin et al. 1997). Activation of tin by twi might be conserved between Drosophila and spiders based on data on twi expression from another spider, A. tepidariorum (Yamazaki et al. 2005). We also recovered a fragment of a twi gene from Cupiennius but were not able to detect expression in embryos via in situ hybridizations (not shown). Based on the conservation of twi expression between Drosophila and Archaearanea (Yamazaki et al. 2005), it is very unlikely that twi is not involved in mesoderm formation of Cupiennius. We therefore assume that there is a second twi gene in Cupiennius that we could not recover in our PCR screen; it is known that several genes in Cupiennius are present as duplicated copies (discussed in Schwager et al. 2007). As we do not have twi data from Cupiennius, we have to rely on the published data on twi in the spider Archaearanea (Yamazaki et al. 2005). The Achaearanea twi gene is expressed in the entire developing mesoderm underlying the ectoderm as segmental stripes, similar as in the insects Drosophila and Tribolium (Handel et al. 2005). The expression of twist seems to be correlated with the formation of coelomic sacs that initially develop as single structures but that split with the beginning ventral fission of the germ band (Wallstabe 1908; Yamazaki et al. 2005). This ventral fission is required for inversion in spiders that results in dorsal closure (Wallstabe 1908; Prpic and Damen 2005). The lateral split of the twist stripes into two patches, one at each side of the embryonic midline, correlates with the ventral fission in Achaearanea (Yamazaki et al. 2005). The early tin expression in the Cupiennius embryo is in dorsal patches in a corresponding position (Fig. 3D). The initial function of twi and tin in mesoderm (and hence heart) development therefore may to be conserved between the fly and the spider. After initial expression in dorsal patches, tin is restricted to the very dorsal edge of the developing embryo that will give rise to the spider heart precursors. This transformation of tin expression from segmental patches (Fig. 3D) into a continuous line (Fig. 2A) might be due to morphological movements of tin expressing mesoderm cells after decomposition of the coelomic sacs into autonomous cell groups. The process of mesoderm realignment has not been described for chelicerates so far, but it may happen in a similar way as described for insects (e.g., Ullmann 1964).
Acknowledgment
We are grateful to Diethard Tautz for continuous support and discussion, Nikola-Michael Prpic and Alistair McGregor for discussion and critical reading of the manuscript, and Manuel Aranda and Henrique Marques-Souza for T. castaneum embryos. This work was supported in part by the Deutsche Forschungsgemeinschaft via Project A12 of the SFB 572 of the Universität zu Köln and by the European Union via the Marie Curie Research and Training Network ZOONET (MRTN-CT-2004-005624).




