Mitochondrial DNA suggests multiple colonizations of central Philippine islands (Boracay, Negros) by the sedentary Philippine bulbul Hypsipetes philippinus guimarasensis (Aves)
Mitochondriale DNA deutet auf multiple Kolonisierungen der zentralen philippinischen Inseln (Boracay, Negros) durch den sedentären Philippinischen BülbülHypsipetes philippinus guimarasensis(Aves) hin
Abstract
enIn this study, we have used fragments of three mitochondrial genes (Control Region, CR; transfer RNA for methionine, tRNA-Met; NADH dehydrogenase subunit 2, ND2 for a total of 1066 bp) to reconstruct the phylogeographic history of the endemic Philippine bulbul (Hypsipetes philippinus) at the scale of the central area of the Philippine archipelago. The study includes two of the five recognized subspecies (guimarasensis and mindorensis), 7 populations and 58 individuals. Multiple phylogenetic and network analyses support the existence of two reciprocally monophyletic maternal lineages corresponding to the two named subspecies. Molecular clock estimates indicate that the split between the two subspecies is consistent with the Pleistocene geological history of the archipelago. Patterns of relationships within guimarasensis are biogeographically less clear. Here, a combination of vicariance and dispersal needs to be invoked to reconcile the molecular data with the geographical origin of samples. In particular, the two islands Boracay and Negros host mitochondrial lineages that do not form monophyletic clusters. Our genetic data suggest multiple independent colonization events for these locations.
Zusammenfassung
deIn dieser Untersuchung haben wir drei Abschnitte mitochondrialer Gene (Kontrollregion, CR; Transfer-RNA für Methionin, tRNA-Met; NADH dehydrogenase Untereinheit 2, ND2, insgesamt 1066bp) analysiert, um die Phylogeographie des endemischen philippinischen Bülbüls (Hypsipetes philippinus) auf den zentralen Inseln der Philippinen zu rekonstruieren. Die Untersuchung umfaßt 2 der fünf beschriebenen Unterarten (guimarasensis und mindorensis), 7 Populationen und 58 Individuen. Multiple phylogenetische und Netzwerk-Analysen unterstützen die Existenz von zwei reziprok monophyletischen maternalen Linien, die den beiden Unterarten entsprechen. Abschätzungen unter Annahme einer molekularen Uhr zeigen eine Übereinstimmung zwischen dem putativen Alter der Unterarten und der pleistozänen Geologie der Philippinen. Innerhalb von guimarasensis sind die biogeographischen Verhältnisse weniger deutlich. Hier muss eine Kombination aus Vikarianz und Dispersion herangezogen werden, um die molekularen Daten mit dem geographischen Ursprung der Proben in Einklang zu bringen. Vor allem die beiden Inseln Boracay und Negros beherbergen mitochondriale Linien, die keine monophyletische Verwandtschaft zeigen. Unsere Daten deuten auf multiple unabhängige Besiedelungen dieser Inseln hin.
Introduction
Ever since Charles Darwin formulated his hypothesis on how the finches inhabiting the Galapagos Islands evolved into a number of different species, oceanic islands have represented a prime choice for evolutionary studies. Islands have traditionally been considered natural laboratories to illustrate evolutionary processes at and above the species level. In the last two decades, thanks to the advent of molecular tools, islands have become the ideal location also to enlighten the genetic changes that take place at the population level in the course of dispersal and gene flow. In particular, depending on the distance among islands and the dispersal abilities of the organism under study, the predictions of general dispersal models (e.g., stepping stone model, continent-island model; cf. Hedrick 2000) can be evaluated.
The Philippines constitute a tropical archipelago made up of more than 7,000 islands, most of which are volcanic in origin. Its complex geological evolution is well known (Hall 1996, 1998, 2001, 2002) and provides a unique frame for evolutionary studies. The archipelago originated following the opening of oceanic basins in the Cretaceous; fragments of these basins were the substratum for the development of volcanic arcs, which in turn gave rise to the Philippines (Tamayo et al. 2004). The archipelago has never had direct connection to the Asian mainland, except for its easternmost portion (Palawan Island). Indeed, Palawan Island does not belong to the oceanic region of the Philippines (which is the focus of this study) and, biogeographically, is part of the Sunda Shelf region (Heaney 1986). During the Pleistocene glaciations, changes in the sea level connected the different islands of the archipelago. In particular, three large aggregate islands had repeatedly being formed (Greater Luzon, Greater Negros-Panay and Greater Mindanao), while other more isolated islands had always remained independent or formed smaller groups (e.g., Mindoro and Semirara, Evans et al. 2003). Evans et al. (2003) termed Greater Luzon, Greater Negros-Panay and Greater Mindanao as ‘Pleistocene aggregate island complex (PAIC)’.
The complex geological history of the archipelago has had a great impact on the evolution of its fauna (Jones and Kennedy 2008; Sheldon et al. 2009). Indeed, we now observe an extremely high rate of endemism (over 57% in the major faunal groups are endemics) with many lineages limited to single islands or PAIC (Oliver and Heaney 1996; Roberts 2006a,b). Because of these characteristics, the Philippine archipelago is considered one of the most species-rich biodiversity hot spot on earth (Myers et al. 2000). Regrettably, this natural treasure is at present increasingly threatened by a dramatic loss of its forest. Forest cover has declined from nearly 100% to 18% in the last 100 years (Oliver and Heaney 1996; ESSC 1999) and moist forests of the Philippines are now classified as the eighth most vulnerable forest ecoregion in the world (WWF 2001). The extant-forested habitat is of critical importance to many groups of organisms, but particularly to birds (Collar et al. 1999).
The object of this study is one the most common birds in the area, the endemic Philippine bulbul Hypsipetes philippinus (Passeriformes, Pycnonotidae). The species is resident and does not migrate seasonally. It is considered omnivorous, although with a clear preference for fruits. The Philippine bulbul thus plays an important role in seed dispersal. Because of its abundance in primary as well as secondary forests, it is among the most important generalist frugivores/seed dispersers in forest habitats; 50% of all net captures of birds in the understory belong to this species (Curio pers. comm.). Consequently, it has a prominent role in promoting the regeneration of clear-felled land (Hamann and Curio 1999). Field observations have demonstrated that individuals rarely fly outside forest canopy (Bennett and Owens 2002). These ecological characteristics, together with the morphological differences among subspecies, possibly indicate a tendency for the species to become locally subdivided. If these assumptions were valid, the Philippine bulbul could represent a further example to elucidate the evolutionary processes driven by insularity. The species range extends across the entire archipelago (Kennedy et al. 2000). Populations living on different islands show slight but constant morphological differences in shade of colour and boldness of shaft streaks on throat; this led to the description of five subspecies (Forster 1795; Steere 1890; Hartert 1916; duPont 1980). Here we focus on one of the five subspecies, H. philippinus guimarasensis. This is the second most distributed subspecies. Its range includes the large islands of Panay and Negros and some of the surrounding islets (i.e., the former Greater Negros-Panay PAIC). It thus represents a good candidate to shed light on processes of diversification within this Pleistocene island complex.
The aim of this study is to produce a phylogeographic hypothesis for H. philippinus guimarasensis at the scale of the Greater Negros-Panay PAIC. To pursue such a goal, we covered most of the subspecies’ range with our sampling. In addition, to place the study in a systematic and phylogenetic context, we analysed one population attributed to the subspecies H. philippinus mindorensis from Semirara. The name of this second subspecies derives from the large island of Mindoro; the subspecies is limited to this island and to the small Semirara Island, which is located approximately halfway between Mindoro and Panay (Fig. 1). Mindoro and Semirara formed a single island during Pleistocene glaciations. We report on variation in the nucleotide sequence of about 1 kb of mitochondrial DNA (mtDNA; three genes) in seven populations and 58 individuals (Fig. 1). We address the issues of how genetically distinct and cohesive are the different populations and how they are related to one another. Our ultimate goal is to understand of how genetic variation is partitioned among populations and islands. Our data will add to those already existing for a few songbirds (Jones and Kennedy 2008) and some other taxonomic groups from the same area (mainly fruit bats and rodents; Jansa et al. 2006; Roberts 2006a,b) to provide a unified framework for the prioritization of conservation efforts in the Philippines.
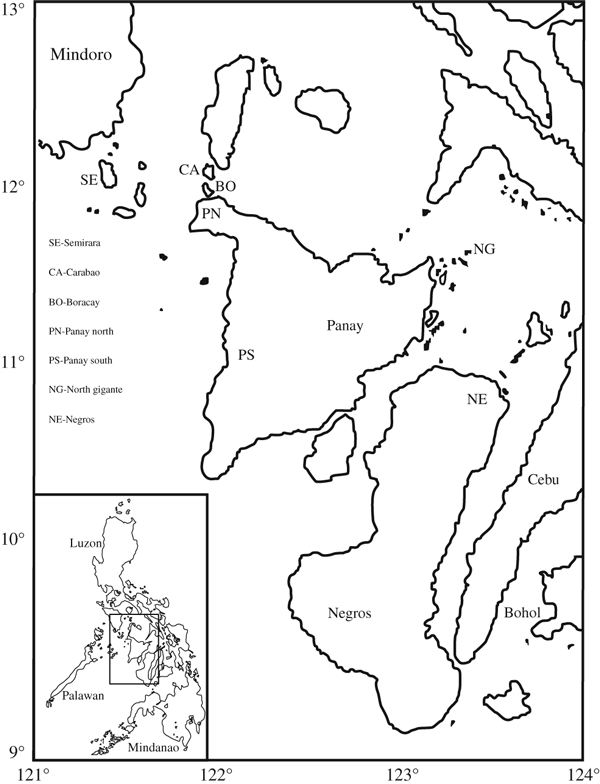
Map of the Philippines showing the locations from which populations of the Philippine bulbul were sampled. The inset shows the entire archipelago and the area considered in the present study. Names of the major islands are also given. Abbreviations are population codes; sample sizes (N) are given in parentheses. Bala Bago, Semirara Is., SE (N = 11); Combot, Carabao Is., CA (N = 11);Yepak, Boracay Is. BO (N = 10); Sibaliw, Panay Is. (North), PN (N = 19); Hamtang, Panay Is. (South), PS (N = 3); Granada, North Gigante Is., NG (N = 1); Patag, Negros Is., NE (N = 3)
Materials and Methods
Sampling
Blood samples were taken from 58 specimens of the Philippine bulbul Hypsipetes philippinus from seven localities following standard procedures (Fig. 1). The 47 specimens from Carabao, Boracay, North Gigante, Panay (both North and South) and Negros belong to the subspecies H. philippinus guimarasensis, while the eleven specimens from Semirara are attributed to the mindorensis subspecies.
DNA amplification and sequencing
Total genomic DNA was extracted from blood samples using the DNeasy kit (Qiagen, Hilden, Germany). We obtained DNA sequences from three mitochondrial genes: control region (CR; 685 bp), transfer RNA for methionine (tRNA-Met; 30 bp) and NADH dehydrogenase subunit 2 (ND2; 351 bp). We used two different primer pairs for all PCR amplifications and sequencing. The primer pair LCR3 and H1248 (Tarr 1995) was used to amplify and sequence the CR fragment, while primer pair L5215 and H5578 (Hackett 1996) was used for the tRNA-Met and ND2 genes. Double-stranded PCRs were performed on a Whatman Biometra thermal cycler (TGradient). We used standard PCR conditions with the following locus-specific annealing temperatures: Ta = 55°C for CR and Ta = 52°C for tRNA-Met and ND2. We purified PCR products with the QIA-quick PCR purification kit (Qiagen, Hilden, Germany). For sequencing we used the BigDye v.3.1 Terminator Cycle sequencing kit (Applied Biosystems, Foster City, CA, USA). The Multiscreen-HV (Millipore, Billerica, MA, USA) purified products were analysed on an AB3100 multicapillary automatic sequencer following manufacturer protocols. Sequences were submitted to GenBank (Accession No. GU214771–GU214796 for CR, GU227784–GU227797 for tRNA-Met and ND2). As an outgroup, we amplified and sequenced the same regions for European Magpie (Pica pica; GU227799 for CR; GU22798 for tRNA-Met and ND2).
Phylogenetic analyses
We aligned sequences using BioEdit version 7.0.0 (Hall 1999). We observed only few indels in the alignments (only in the CR). These were therefore straightforward for all genes. We analysed data phylogenetically by Neighbor-Joining (NJ) (Saitou and Nei 1987) and Bayesian methods (Rannala and Yang 1996; Mau and Newton 1997; Larget and Simon 1999; Mau et al. 1999; Huelsenbeck and Ronquist 2001) to derive a haplotype tree. Gene fragments were analysed both separately and combined. Phylogenetic searches run on each gene separately were identical to that based on the combined data set. We therefore present and refer only to the latter in the remainder of the paper. We used the Akaike Information Criterion (AIC) implemented in MODELTEST (Posada and Crandall 1998) to determine the best model of sequence evolution. We used the output of MODELTEST to calculate distances for NJ analyses. For the Bayesian approach, we employed the same models of sequence evolution allowing site-specific rate variation partitioned by gene and, for ND2, by codon positions. MRBAYES was run for 2 million generations with a sampling frequency of 100 generations. We ran one cold and three heated Markov chains. To establish if the Markov chains had reached stationarity, we plotted the likelihood scores of sampled trees against generation time. Trees generated before the stationarity phase were discarded as ‘burn-in’ (first 10% of the sampled trees), and posterior probability values for each node were calculated based on the remaining 90% of sampled trees. These trees were used to construct a 50% majority rule consensus tree using PAUP* 4.0β10 (Swofford 2003). The robustness of the NJ phylogenetic hypotheses was tested by 1000 bootstrap replicates (Felsenstein 1985). We used the Eurasian magpie (Pica pica; order Passeriformes, family Corvidae) and the domestic chicken (Gallus gallus; order Galliformes, family Phasianidae, GenBank Accession No AP003318) as outgroups in the phylogenetic analyses. These outgroups were chosen for the combined data set because of the lack of availability of sequences for all three analysed genes for any other bulbul species. Note that a phylogenetic search based on ND2 only, using the bulbul Pycnonotus goavier (DQ402237) as an outgroup, yielded the same topology (data not shown).
We took advantage of a recent paper on bulbul phylogeny (Moyle and Marks 2006). Based on these data, we calculated inter-specific uncorrected-p genetic distances within nine bulbul genera (Pycnonotus, Andropadus, Phyllastrphus, Xanthomixis, Bleda, Criniger, Alophoixus, Ixos and Nicator) at the ND2 level. We then used these data as a background for the level of genetic divergence we found between the two subspecies of the Philippine bulbul we included in the study and to discuss the possible taxonomic implications of our findings.
The molecular clock hypothesis was tested via a relative-rate test as implemented in the program RRTree (Robinson-Rechavi and Huchon 2000). For the test, we sorted haplotypes by subspecies and we used the 2-parameter distance of Kimura (Kimura 1980). Time estimates were calculated by using the formula T = D/2 r, where D is the uncorrected-p genetic distance and r is the mutation rate. We used the passerine mtDNA rate (2–2.4% substitutions per million years; myr) proposed by Tarr and Fleischer (1993) for the Hawaiian honeycreepers.
We also constructed a haplotype network under the statistical parsimony procedure (Templeton et al. 1992) implemented in TCS (Clement et al. 2000); different haplotypes were linked only if they had a 99% probability of being justified by the parsimony criterion. In this analysis, gaps were treated as a fifth state.
Population genetic analyses
Analysis of molecular variance (amova) was performed using Arlequin ver 3.01 (Excoffier et al. 2005), with populations grouped into recognized subspecies (Steere 1890). This approach produces three variance estimates: among subspecies, among populations within subspecies and within populations. Significance of variance estimates was obtained using a randomization procedure (1000 permutations). We estimated Φ-statistics, which are haplotypic correlation measures analogous to F-statistics. The Φ-statistics are defined as follows: ΦCT is the correlation of random haplotypes within subspecies relative to that of random haplotypes drawn from all populations; ΦSC is the correlation of random haplotypes within populations relative to that of random haplotypes drawn from the subspecies to which that population belongs; and ΦST is the correlation of random haplotypes within populations relative to that of random haplotypes drawn from the whole (Excoffier et al. 1992). The input matrix for the amova analyses was obtained by calculating the number of differences between unique haplotypes in pairwise comparisons. Pairwise FST values for all pairs of populations were also calculated using Arlequin 3.01 (Excoffier et al. 2005) and their significance assessed by 1000 permutations with Bonferroni correction. We used the Mantel test as implemented in Arlequin 3.01 (Excoffier et al. 2005) to test for a correlation between matrices of pairwise FST values and geographical distances (in km) among populations. A positive and significant correlation between the two matrices would indicate an isolation-by-distance pattern in the data (Slatkin 1993).
Arlequin 3.01 (Excoffier et al. 2005) was finally used to carry out multiple mismatch analyses to compare the demographic history of each population/clade and to test for sudden population expansions. After having checked the results of the phylogenetic analyses, we conducted the mismatch analyses on the following hierarchical levels: each population separately, all populations excluding Semirara, all populations excluding Semirara and those haplotypes from Boracay which were basal and monophyletic in the phylogenetic trees, only the latter group of haplotypes from Boracay, and all populations simultaneously. For each of these groups, we computed the moment estimator of time to the expansion (τ), and the mutation parameters before (θ0 = 2μ N0) and after (θ1 = 2μ N1) the expansion using a parametric bootstrap approach (1000 simulations), where μ is the mutational rate for the whole haplotype and N is the female effective population size. We also computed the raggedness index of Harpending under the sudden expansion model (Harpending 1994) and used the sum of square deviations (SSD) between the observed and the expected mismatch as a test statistic for the validity of the estimated stepwise expansion model (Schneider and Excoffier 1999). Finally, we used the equation τ = 2 μt to infer a time-scale for the demographic expansion (where t is the expansion time in generations). We assumed an average generation time of 1 year (Kennedy et al. 2000), and the same substitution rate we adopted for the molecular clock calculations.
Results
Sequence variation, haplotype distribution and measures of molecular diversity
We sequenced 1066 bp of mtDNA (685 bp for CR, 30 bp for tRNA-Met and 351 bp ND2) for each of the 58 Philippine bulbuls. We found only few indels in the alignment; all these indels were in the CR and most of them were between the outgroups and the ingroup. Level of sequence variation was slightly higher in ND2 than in CR. 12.53% of the ND2 sites were variable with 11.96% of them being also parsimony informative. CR had 9.63% of variable sites and 8.61% of parsimony informative sites. As expected, we found most of the variation in ND2 3rd codon positions (29.05% of variable sites; 28.2% of parsimony informative sites). We found no variation in tRNA-Met. In the combined data set, 10.31% of the sites were polymorphic and 9.47% of them were also parsimony informative. Sequences were generally A + T rich (A + T percentages ranged from 39.6% in ND2 3rd codon positions to 58.0% in tRNA-Met) and anti-G biased (12.9–15.3% in tRNA and ND2 3rd codon positions, respectively). Chi-square tests of homogeneity of base frequencies among taxa were always not significant, independently from the data partition tested (all genes together, each gene separately, 1st, 2nd and 3rd ND2 codon positions separately).
The 58 sequences we obtained for this study defined a total of 35 distinct haplotypes (see supplementary table S1). The number of haplotypes in those populations from which more than one individual was sampled ranges from three (Panay South and Negros) to 11 (Panay North). Only three haplotypes (8, 9 and 23) are shared among different locations. Indeed, we found haplotypes 8 and 9 in individuals from Panay North, Panay South, and Carabao and from Panay North and Carabao, respectively. Haplotype 23 is shared between Panay North and South. All remaining haplotypes are unique to single locations (see supplementary table S2). The population sampled on Negros shows the highest amount of genetic variability while Carabao has the lowest.
Phylogenetic and network analyses
Figure 2 shows the NJ tree based on HKY+ Γ distances (Ti/Tv ratio = 1.467; base frequencies A = 0.27, C = 0.29, G = 0.14, T = 0.30; variable rates among sites with shape parameter α = 0.007; parameters for the model selected by MODELTEST) and summarizes the result of the Bayesian searches. Either method produced trees with very similar topologies. Within the ingroup, haplotypes of individuals formally attributed to the two subspecies (guimarasensis and mindorensis) cluster in reciprocally monophyletic clades. Both clades had maximum statistical supports in NJ and Bayesian searches. Within the guimarasensis clade three haplotypes (1, 3 and 5) found in the population sampled on Boracay form a strongly supported monophyletic group; three additional haplotypes (2, 7 and 4) from this location cluster together with good statistical support but are nested within haplotypes found in populations from other islands. No other group of haplotypes received statistical support in the phylogenetic searches.
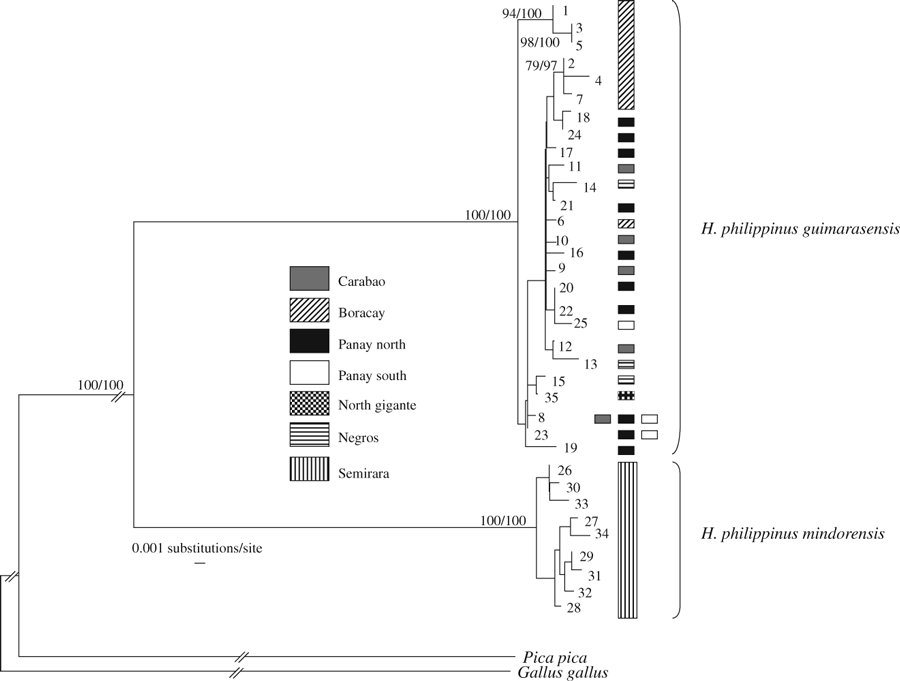
MtDNA haplotype tree obtained by the Neighbor-Joining (NJ) method under the HKY+Γ model of sequence evolution. Only statistical supports ≥75% for both the NJ (1000 bootstrap replicates) and Bayesian searches (2 000 000 generations) are reported on branches. Branch lengths are proportional to the amount of genetic divergence within the Philippine bulbul clade only; for the outgroup species branch lengths have been bracketed. Numbers for haplotypes are as in Supplementary Table S1; different shadings identify the geographical origin of samples and match those in Fig. 1
The relative-rate test did not reject the molecular clock hypothesis between the two subspecies (p = 0.949), suggesting that the variation in substitution rate among them is negligible. Given the degree of genetic divergence observed between the two subspecies (uncorrected-p value = 0.099 ± 0.003) and by using the passerine molecular clock rate of Tarr and Fleischer (1993), we estimate that the split between them ranges in age from 1.95 ± 0.025 myr to 1.63 ± 0.02 myr.
Mean uncorrected-p genetic distance values based on ND2 sequences (calculation based on data from Moyle and Marks 2006) for five genera and 28 species of bulbul are summarized in Table 1. Divergence at the species level ranges between 0.103 ± 0.075 within the genus Criniger (four species) to 0.183 (genus Ixos; two species). The inter-specific mean uncorrected-p value averaged over genera is 0.146 ± 0.025.
| Genera | No of species | Mean uncorrected-p ± SD |
|---|---|---|
| Alophoixus | 3 | 0.163 ± 0.032 |
| Criniger | 4 | 0.103 ± 0.075 |
| Ixos | 2 | 0.183 |
| Pycnonotus | 13 | 0.175 ± 0.023 |
| Xanthomixis | 4 | 0.123 ± 0.032 |
Figure 3 shows the haplotype network obtained via the statistical parsimony procedure implemented in TCS (Clement et al. 2000). Similarly to what we observed in the phylogenetic searches, the network analysis identifies two major groups of haplotypes, which correspond to nominal subspecies. Within the guimarasensis group, there is little structuring by geographical origin of samples. There are two major groups of haplotypes from Boracay, which are identical to those in the NJ and Bayesian trees (haplotypes 1, 3, 5 and haplotypes 2, 4, 7, respectively). These two groups are respectively linked to haplotypes 8 (shared among Carabao, Panay North and Panay South) and 9 (in common between Carabao and Panay North). The single individual from North Gigante is nested between haplotype 8 and haplotype 15 from Negros.
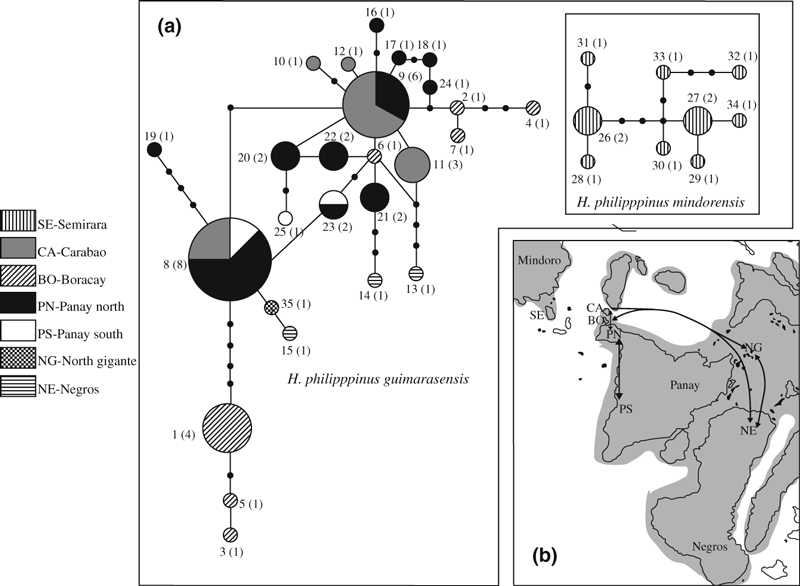
A) Haplotype networks derived from 1066 bp of mtDNA (control region, tRNA-met and ND2). The relative size of the circles is proportional to the number of individuals carrying that particular haplotype; pie slices indicate the fraction of each population contained within each haplotype. Different shadings identify the geographical origin of samples (see Fig. 1). The numbers close to each circle identify the haplotypes and how many individuals carried that particular haplotype (in parentheses). Black dots are missing haplotypes. Note that haplotypes from Semirara (H. p. mindorensis) are between 79 and 90 mutational steps away from those found elsewhere. B) Schematic reconstruction of the phylogeographic history of the Philippine bulbul. Shaded areas indicate the Greater Panay-Negros Pleistocene aggregate island complex and the aggregate Mindoro and Semirara, as they would have been during the Pleistocene. Solid arrows indicate ongoing gene flow. Double arrows indicate inferred back migrations between locations while single arrows suggest unidirectional gene flow. Dashed lines identify historical events of gene exchanges among locations. Numbers indicate approximate temporal order. See Discussion for details
Population genetics
Table 2 reports the results of the hierarchical analysis of molecular variance (amova). This analysis shows that 94.04% of the variation is because of differences between subspecies, 1.27% among populations within subspecies and 4.65% within populations. The global ΦST is 0.95 (p < 0.001). The fixation index among populations within subspecies (ΦSC) is 0.21; (p < 0.001). This indicates a substantial population structuring within the two subspecies, particularly in H. philippinus guimarasensis for which a representative number of populations were analysed.
| Source of variance | Sum of squares (df) | Variance component | p | Fixation index | % of variance |
|---|---|---|---|---|---|
| Among subspecies | 714.834 (1) | Va = 39.6477 | <0.001 | ΦCT = 0.94 | 94.04 |
| Among pops. within subspecies | 28.053 (5) | Vb = 0.5338 | <0.001 | ΦSC = 0.21 | 1.27 |
| Within populations | 99.837 (51) | Vc = 1.9576 | <0.001 | ΦST = 0.95 | 4.65 |
Pairwise FST values are presented in Table 3. For this analysis, the two populations from Panay have been pooled together as we found no genetic differentiation between them (FST = −0.060). The FST values between the two subspecies are high and significant in all cases. Within H. philippinus guimarasensis, few comparisons yielded significant FST values; these were always substantially lower than the values found between the two subspecies. The Mantel test revealed a non-significant (p = 0.335) association between geographical distances among populations and pairwise FST values.
| Population | SE | CA | BO | PA | NE |
|---|---|---|---|---|---|
| SE | – | 62 | 63 | 74 | 259 |
| CA | 0.964 | – | 10 | 98 | 212 |
| BO | 0.937 | 0.353 | – | 22 | 209 |
| PA | 0.957 | 0.072 | 0.308 | – | 187 |
| NE | 0.945 | 0.393 | 0.280 | 0.268 | – |
Table 4 and Figure 4 present the results of the mismatch distribution analyses conducted at different hierarchical levels. The mismatch distributions differed substantially among populations and clades. Haplotypes 1, 3 and 5 from Boracay (see Fig. 2) had the lowest mean number of differences (observed mean = 1.400); this value is almost five times lower than the observed mean (6.533) we obtained when all haplotypes from Boracay are analysed simultaneously (i.e., independently from the results of our phylogenetic analyses). A model of sudden population expansion was supported in most cases but four (Panay North, Panay South, Negros and all populations excluding haplotypes 1, 3 and 5 from Boracay). Graphs presented in Fig. 4 reveal that mismatch distributions vary from unimodal to bimodal (Panay South and all populations either with or without excluding haplotypes 1, 3 and 5 from Boracay) to multimodal (all haplotypes found on Boracay). The mismatch distribution including all samples has two clear peaks, with one peak corresponding to differences between subspecies and the second peak corresponding to differences within subspecies. Table 4 also shows the timing of the most important demographic expansions in the different lineages. Time estimates range in age between 35.6 and 270.2 kyr. We calculated time estimates only for those lineages where a sudden population expansion was supported by the data.
| Population | τ | Obs. mean | θ0 | θ1 | p(HARP) | p(SSD) | t expansion (kyr) |
|---|---|---|---|---|---|---|---|
| SE | 5.090 (2.18–6.66) | 4.655 | 0.001 (0.00–4.33) | 87.319 (9.90–131.69) | 0.250 | 0.400 | 119.3 (51.1–156.1) |
| CA | 1.855 (0.71–3.58) | 1.636 | 0.000 (0.00–0.93) | 51.797 (1.22–69.94) | 0.500 | 0.750 | 43.5 (16.6–83.9) |
| BO | 11.525 (5.13–19.07) | 6.533 | 0.000 (0.00–0.00) | 10.945 (6.96–212.04) | 0.450 | 0.600 | 270.2 (120.3–447.2) |
| PN | 3.441 (1.51–4.45) | 3.146 | 0.001 (0.00–1.43) | 55.098 (2.59–167.01) | 0.050 | 0.050 | 80.6 (35.4–104.3) |
| PS | 6.288 (3.41–22.26) | 4.000 | 0.000 (0.00–15.15) | 34.761 (2.18–181.40) | 1.00 | <0.05 | – |
| NE | 7.329 | 6.667 | a | a | <0.05 | <0.05 | – |
| BOb | 3.279 (1.96–15.22) | 1.400 | 0.000 (0.00–11.43) | 2.476 (0.29–8154.98) | 0.700 | 0.800 | 76.8 (45.9–356.9) |
| All populations without SE | 2.119 (2.35–9.52) | 4.264 | 2.605 (0.00–2.59) | 27.603 (10.83–48.08) | 0.600 | 0.600 | 56.9 (55.1–223.2) |
| All populations without BOb | 3.642 (2.23–5.36) | 1.982 | 3.303 (0.00–10.85) | 42.43 (11.32–361.96) | 0.750 | 0.050 | 85.4 (52.2–125.7) |
| All populations | 1.522 (0.00–17.45) | 29.281 | 5.263 (0.00–11.39) | 37.861 (7.50–100.95) | 0.550 | 0.850 | 35.6 (0–409.2) |
- aLeast square procedure to fit model mismatch distribution and observed distribution did not converge after 1800 steps. bonly haplotypes 1, 3 and 5 from Boracay.
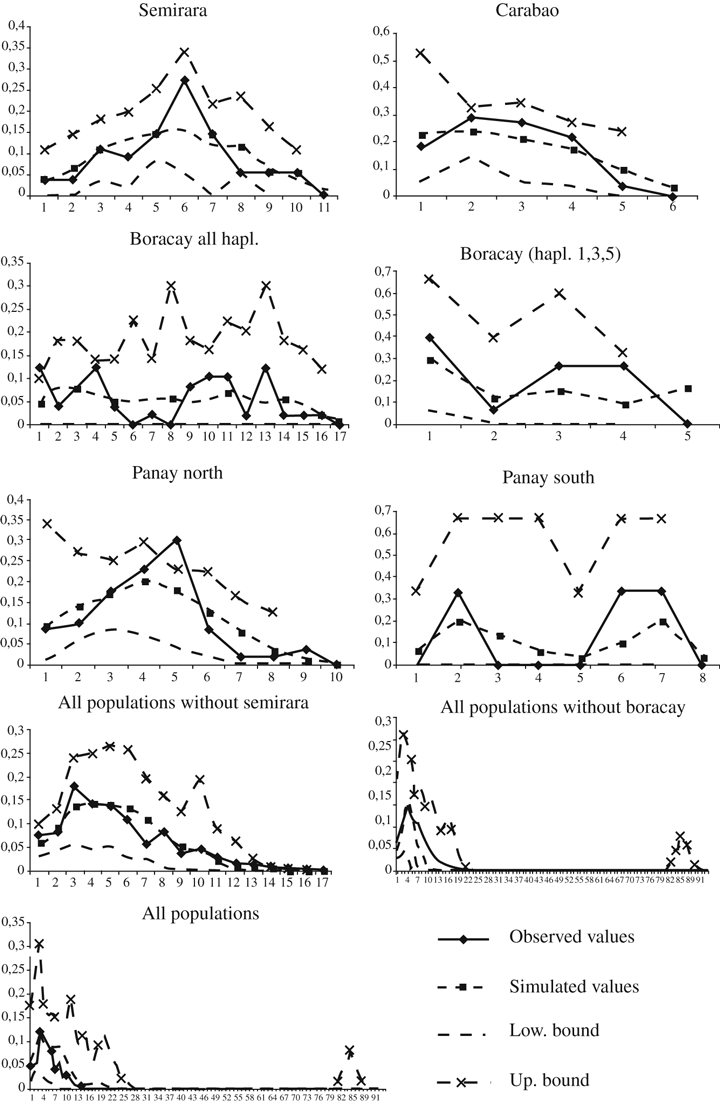
Frequency distribution of sequence differences for all possible pairs of individuals in the populations of the Philippine bulbul included in the study obtained by mismatch analyses. Populations have been analysed separately and pooled together; mismatch distributions have been also calculated for all populations excluding Semirara and for all populations excluding haplotypes 1, 3 and 5 from Boracay. The x-axis shows the actual number of mutational differences; the y-axis depicts relative numbers of pairwise comparisons. Observed values, values of the model fitted to the data and the 2.5 and 97.5 percentile values (1000 simulations) are shown in the graphs (see captions for different symbols)
Discussion
Molecular systematics
Although Philippine bulbul systematics was not the purview of the present study, we shortly discuss implications of our data as a starting point for future researches on the species taxonomy. Our data have revealed the existence of two major geographically distinct and reciprocally monophyletic maternal lineages (Table S1; 2, 3). Of these two lineages, one corresponds to the subspecies mindorensis on Semirara; the second is represented by the subspecies guimarasensis from Carabao, Boracay, Panay, North Gigante and Negros. MtDNA data are thus in agreement with previous taxonomic studies (Forster 1795; Steere 1890; Hartert 1916; duPont 1980). These authors distinguished five subspecies based on a combination of differences in the plumage pattern (i.e., shaft streaks on throat) as well as in songs. Among the five subspecies, mindorensis has the most distinct plumage with very inconspicuous streaks on the throat and also sings the most different song. The two subspecies we included in the study are separated by a mean genetic distance (uncorrected-p; ND2 gene only) of 0.099 ± 0.003. This value is always lower than the average inter-specific genetic distances we obtained by re-analysing the ND2 data set of Moyle and Marks (2006), which includes 28 morphologically well-differentiated bulbul species (Table 1). Whether the level of genetic divergence we found is indicative of a speciation process already completed or still ongoing is difficult to tell. Nevertheless, there are several lines of reasoning to support the idea that the two subspecies do represent truly independent lineages. The most striking one is that we observe complete reciprocal monophyly between them. This implies that gene flow has been absent for a considerable lapse of time to result in coalescence of lineages on Semirara on one hand and on Panay, Negros and surrounding islets on the other. This hypothesis also makes clear sense biogeographically. Additionally, the amova results demonstrate that the vast majority of the detected genetic heterogeneity is because of differences between the two subspecies. Finally, we obtained large and highly significant pairwise FST values between Semirara and all other locations. It is also true that this scenario might be challenged by the fact that we sequenced only maternally inherited genes. This would represent a severe bias in the case of a strong male-biased dispersal, which could potentially homogenize gene frequencies at nuclear loci. At the moment, there are no sufficient ecological/field data to accept or disregard the male-biased dispersal hypothesis, and complementary genetic data from nuclear loci are not yet available. It must be noted, however, that the degree of mtDNA geographical structure and the length of the branches separating the two subspecies in the haplotype tree (Fig. 2) suggest a long history in the absence of gene flow and are hardly reconcilable with a sex-biased dispersal scenario.
We are aware that our taxon sampling is not adequate to draw definitive conclusions on the systematics of the species. To address this issue properly, we would have needed a denser sampling throughout the entire Philippines.
Phylogeography and population genetics
The deep phylogeographic break between the two subspecies is very likely related to the geological history of the Philippines, which has been described in detail (see Introduction). Our samples come from the former (i.e., Pleistocene) Greater Negros-Panay PAIC and aggregate Mindoro and Semirara. Our time estimates place the split between guimarasensis and mindorensis at 1.63–1.95 myr. These dates correspond to the Biber/Donau interglacial. Our time estimates need to be considered cautiously because they are based on a single mitochondrial marker and therefore most likely associated with a high variance. Still, they are in good agreement with the geological evolution of the archipelago. Although Greater Negros-Panay and the aggregate Mindoro and Semirara were not joined by a land bridge during glaciations, we hypothesize that some gene flow might have taken place because of over-water movements. These episodes of gene flow were probably favoured by the much closer proximity of coastlines during glaciations, when the sea level dropped to 120 m below the current level (Fig. 3B, Heaney 1986). During interglacial times, with the rising of sea level, coastlines had almost the same shape as they have nowadays. This presumably led to a strong reduction (if not a complete cessation) of gene flow between the two landmasses. We have to emphasize that this is just a preliminary attempt to reconstruct the among-PAICs relationships in the species. In particular, we stress here that, because of our limited sampling across the archipelago, a sister taxa relationship between the two subspecies should not be taken for granted. The inclusion in the analysis of all other subspecies is indispensable to reconstruct the timing and pattern of diversification of the species at the scale of the entire archipelago. Partial coincidental support to our findings comes from a recent mtDNA study (based on cytochrome b and ND2 genes) by Roberts (2006a) on the endemic Philippine fruit bat Haplonycteris fischeri. The sampling for that study covered all recognized PAICs. Similarly to what we obtained, this author found a monophyletic maternal lineage on Mindoro. Interestingly, this lineage was sister to another monophyletic clade restricted to Panay, Negros and Luzon. We sampled the former two islands but not the latter, which hosts the nominal subspecies (philippinus).
Our sampling efforts concentrated on the subspecies guimarasensis, for which we covered most of its distribution range. Figure 3B presents a schematic reconstruction of the phylogeographic history of this taxon. The reconstruction summarizes the results of phylogenetic, network and pairwise FST analyses: Gene flow events were inferred from the phylogenetic network; when the respective populations were not differentiated from one another (i.e., non-significant FST), this was considered current gene flow (arrows). Even with these simplifications, it is still evident that there is not a direct link between evolutionary relationships among haplotypes and their geographical origin. Granted that the split between the two subspecies (whether or not these are each other’s closest relatives in the species’ phylogeny) predated the differentiation within them, genetic data cannot safely identify the timing of the different dispersal events within guimarasensis, because of a lack of statistical support in the haplotype tree of Fig. 2. Indeed, there are only two statistically supported nodes in the guimarasensis clade; both of them group haplotypes found on Boracay. Phylogenetic and network analyses (2, 3) suggest that Boracay was colonized at least two times independently. The first colonization of Boracay presumably occurred early in the history of the taxon, as suggested by the basal position of haplotypes 1, 3 and 5 (Fig. 2). On the contrary, the remaining haplotypes found on Boracay (2, 4, 6 and 7) are deeply nested within haplotypes from other locations. This islet shares no haplotypes with any of the other sampling sites. This was quite unexpected given its geographical proximity to both Panay and Carabao. On the other hand, the presence of the same haplotypes on Panay North and Carabao suggests migrations in either direction between these islands. It might well be that haplotypes shared between Boracay and the other populations we analyzsd also exist (or existed in the past) but have not appeared in our sample. There are neither obvious ecological nor geological evidences to account for the degree of genetic isolation of Boracay from the other locations within the same PAIC. Given the data and the vagaries in interpreting our results with regard to this islet, we tentatively propose the following scenario: a first unidirectional migration event brought the ancestor of haplotypes 1, 3 and 5 to Boracay. Later on, a similar event occurred for the ancestor of haplotypes 2, 4, 6 and 7. In both circumstances, genetic data cannot identify whether the colonization of Boracay started from Panay or Carabao. However, the network analysis of Fig. 3A suggests Panay rather than Carabao as the most likely source of migrants to Boracay. This hypothesis is based on the consideration that Panay has three haplotypes (9, 21 and 22) that are only one mutation step away from haplotype 6 from Boracay, while Carabao has only one (9). This scenario would also fit with island biogeography theory, which predicts that large islands act as a source for dispersal of species/individuals to smaller islands (MacArthur and Wilson 1963, 1967).
Relationships among Negros, North Gigante and Panay are also complex. Figs 3A and B suggest that multiple colonization events need to be invoked to reconcile our molecular data with the geographical origin of samples for those areas. Two of the three haplotypes (13 and 14) we found on Negros are independently linked through 2–3 missing haplotypes to haplotypes unique to Carabao and Panay North. The third haplotype (15) shows a direct connection to haplotype 35 found on North Gigante. The latter is in turn only one mutation step away from haplotype 8, which is shared among Panay North, Panay South and Carabao. All this implies that Negros was colonized at least three times independently from Carabao, from Panay North and/or from North Gigante. This scenario also implies that the population on North Gigante presumably originated from a geographically widespread lineage. Unfortunately, we were able to sample only a single individual on North Gigante and this obviously prevents an in-depth test of this hypothesis. Interestingly, a recent study on three song bird species from PAIC also inferred multiple colonization and the lack of a direct link between populations from Panay and Negros, despite of their geographical proximity and their pleistocene connection (Jones and Kennedy 2008).
The failure in detecting an isolation-by-distance pattern in the data suggests that factors other than geographical distance among locations are (or had been) important in shaping the pattern of genetic variation in the subspecies. The Philippine bulbul is strictly bound to forests and tends to avoid flying out from beneath forest canopy. Moreover, a dense understory is particularly critical for a successful nesting (Kennedy et al. 2000). Consequently, variations in forest cover among and within islands as well as changes in the density, composition and persistence of understory are among the most important factors affecting its dispersal potential. Understanding the relative influence of each of these variables would be of paramount importance to better explain levels of genetic connectedness among populations. A recent study aimed to produce a comparative phylogeography for three widespread Philippine fruit bats yielded similar conclusions (Roberts 2006b).
A closer look at the two networks of Fig. 3A reveals some differences in the respective patterns. The mindorensis haplotypes have a clear structure with two major groups and no reticulation between them. Conversely, there are two haplotypes (8 and 9) at high frequency in the guimarasensis data set surrounded by a number of haplotypes carried by single (or very few) individuals. The latter arrangement closely resembles a star-like pattern, which is often indicative of a sudden population expansion. Indeed, most of the groups of haplotypes we tested for such an event (either based on geographical or phylogenetic criteria) showed signs of quick changes in their past demography (Table 4 and Fig. 4). When all populations are analysed simultaneously, the mismatch distribution curve is clearly bimodal and accurately reflects the presence of two different lineages (i.e., the two subspecies) in the area under study. The mismatch distribution is strongly unimodal in mindorensis (a single expansion event dated at about 120 kyr), while graphs for the different groups of guimarasensis show strikingly different patterns. This holds particularly true for Carabao, Boracay and Panay North, in spite of their geographical proximity. The mismatch distribution is smooth for Carabao, jagged for Boracay and strongly unimodal for Panay North. These results suggest that population size has been more stable on Boracay than it has been on either Panay North or Carabao. At first glance, this is partially at odds with our hypothesis of Panay as the most probable source of colonization of Boracay and Carabao. However, to reconcile these views, one has to consider that the demographic changes detected by mismatch distribution analyses all occurred quite recently and in a very short time span (from 85.4 to 43.5 kyr; Table 4). This scenario has two main implications: first, time since divergence has been very likely insufficient to allow for complete sorting of lineages. Second, what looks at a first glance as a very untidy pattern of relationships might well conceal a number of secondary contacts among lineages. During glaciations not only many of the islands that constitute the Philippines were merged in large aggregates but also tropical forests expanded considerably (McKenna and Farrell 2006). This enhanced dispersal and subsequent secondary contacts of lineages that were probably geographically restricted during interglacial times. This hypothesis would explain the lack of resolution in our phylogenetic searches and the failure in detecting a pattern of isolation-by-distance in the data.
Conclusions
Our analyses proved to be informative to shed light on the phylogeography and population genetics of the Philippine bulbul in the central area of the Philippines. Although not exhaustive with regard to the distribution range of the entire species, they show that the genetic pattern of diversification in this forest species is complex. Vicariance needs to be invoked to interpret the timing and pattern of differentiation between the two subspecies. Our data are in remarkable agreement with the available reconstructions of the early Pleistocene geology of the archipelago. At a more shallow taxonomic level (i.e., within subspecies), a combination of vicariance and dispersal is needed to reconcile molecular relationships among haplotypes and their geographical distribution. Compared to other archipelagos like the Galápagos (e.g., Parent et al. 2008), multiple dispersal/colonization across islands appears more common in the Philippines.
The data support the idea that (linear) geographical distance among populations is not the only force driving the evolution of the species. Rather, in a more realistic and modern view, a number of historical (current versus past coastlines) and ecological (forest cover) factors have to be taken into account. Therefore, phylogeographic patterns are the resultant of the interaction of both long- and short-terms processes and do not simply reflect current conditions. All this might have important implications, when it comes to prioritize conservation efforts in a place of imperilled biodiversity like the Philippines.
Acknowledgements
A. Silva-Iturriza was supported by DAAD-FUNDAYACUCHO scholarship. The help by the Philippine Endemic Species Conservation Project (PESCP) was pivotal in providing samples from Panay, Negros and Boracay. We wish to express our gratitude to Prof. Dr. E. Curio for critical advices throughout the different phases of the study. B. Tacud and other project staff of PESCP’s Research Station Sibaliw rendered indispensable assistance in the field. G. Ledesma and his Negros Forest and Ecological Foundation provided logistical help. Financial support is acknowledged from the University of Potsdam. We also thank S. Pfautsch and K. Moll for technical assistance in the lab.




