Population structure, colonization processes and barriers for dispersal in Polish common hamsters (Cricetus cricetus)
Populationsstruktur, Besiedlungsprozess und Ausbreitungsbarrieren des Feldhamsters (Cricetus cricetus) in Polen
Agata Banaszek ([email protected])
Katarzyna A. Jadwiszczak ([email protected]), Mirosław Ratkiewicz ([email protected]), Joanna Ziomek ([email protected]), Karsten Neumann ([email protected])
Abstract
enThe phylogeographic relationships of common hamster (Cricetus cricetus) populations in Poland were determined by the analysis of three partial mtDNA sequences: control region, cytochrome b and 16S rRNA. A phylogenetic tree as well as parsimony network, consistently separate Polish common hamsters into two groups: E1 being so far specific for the area of Poland, and P3 which clusters inside a Pannonian lineage, previously described from the Carpathian Basin. Polish hamsters do not share any haplotypes with the ‘North’– lineage from Germany and Western Europe, although Poland most likely represents the main migration corridor from the eastern distribution centre to the western boundary of the species range. Fossil and DNA data indicate a very recent appearance of the E1 lineage in the Polish Uplands, probably at the very end of the last glaciation. On the other hand, the Pannonian group entered southern Poland as early as the second stadial of the last glaciation (Middle Vistulian 53.35 ka). The hamster lineages in Poland seem to show different population structures and demographic histories.
Zusammenfassung
esDie phylogeographische Beziehung rezenter Populationen des Feldhamsters (Cricetus cricetus) in Polen wurde anhand der Haplotypenverteilung von drei partiellen mitochondrialen Genen (Kontrollregion, Cytochrom b und 16S rRNA) untersucht. Genetische Stammbäume und Netzwerke zeigen, dass polnische Feldhamster in zwei phylogenetische Linien aufspalten, E1 und P3. Die E1-Linie ist bisher nur für Polen bekannt, während die P3-Gruppe zur pannonischen Linie gehört, die bisher nur für das Karpatenbecken beschrieben ist. Polnische Feldhamster teilen keine mitochondrialen Haplotypen mit der ‘North’-Linie in Deutschland und Westeuropa, obwohl Polen als wahrscheinlicher Besiedlungskorridor vom östlichen Verbreitungsschwerpunkt in das westliche Areal infrage kommt. Fossildaten und DNA-Analysen weisen auf eine späte Besiedlung Polens durch die E1-Linie hin, die wahrscheinlich gegen Ende der letzten Eiszeit erfolgte. Im Gegensatz dazu erreichte die pannonische Gruppe Südpolen im zweiten Stadial der Weichseleiszeit. Die beiden Feldhamsterlinien in Polen haben nicht nur verschiedene Populationsstrukturen, sondern auch unterschiedliche demographische Hintergründe.
Introduction
Distribution and diversity of organisms in temperate regions was largely shaped by climatic oscillations during the Pleistocene. The spatial pattern of distinct genetic clades, which has been observed in many species is traditionally explained by allopatric differentiation in isolated glacial refugia (Hewitt 1999). Patterns of late or postglacial re-colonization events can usually be elucidated from the present day distribution of phylogeographical lineages and major geographical or ecological barriers for dispersal can usually be detected. The process of diversification as a consequence of the interplay between expansion from isolated refugia and the restriction of gene flow by ecological and geographical barriers is most easily observed in species with particular ecological demands. The common hamster, Cricetus cricetus L. (1758) is such a species representing the only widespread steppe mammal in central and western Europe. Its European range extends belt-like between approximately 45°–55°N (in European Russia 60°N) from Eastern Europe to some isolated populations in Belgium, the Netherlands and France in the west (Mitchell-Jones et al. 1999).
The Carpathians provide the most important geographic barrier in central Europe dividing common hamsters into two main genetic lineages (Neumann et al. 2005). A northern lineage comprises populations from Germany and adjacent western European countries. A second Pannonian lineage is distributed inside the Carpathian basin (Fig. 1). More phylogroups most probably exist in the eastern main distribution area, however, they have not been defined so far because of the scarcity of data (Neumann et al. 2005). Separation times of 85–147 ka between the two main central European lineages imply that the re-colonization of Europe started from isolated refugia most likely in the southern Russian and Ukrainian steppe zones. Poland must have provided the corridor for range extensions into Germany and western Europe. However, the mitochondrial haplotype of a single hamster from Poland did not provide the expected link between Russian and German hamsters (Neumann et al. 2005). It was thus concluded that the northern distribution range was highly unstable and experienced a number of successive and probably short-lived colonization events from different eastern source populations (Neumann et al. 2005). Moreover, the analysis of the mtDNA control region of Polish hamsters did not resolve the phylogeographic relationships of the populations because of low diversity. The distribution of the haplotypes was geographically structured and it could only be concluded that the area of Poland was most probably colonized from different source populations (Banaszek et al. 2009).
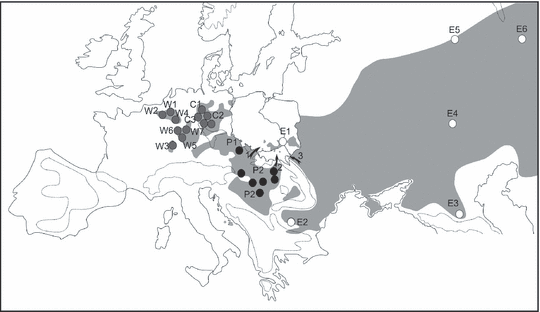
The distribution of the common hamster (Cricetus cricetus L.) phylogroups in Central Europe after Neumann et al. (2005), modified: grey circles – the ‘North’ lineage, black circles –‘Pannonia’, open circles – East samples. The arrows indicate postulated northwards migration routes of the ‘Pannonia’ phylogroup around and through the Carpathians into the area of Poland: 1 – the Moravian Gate, 2 – the Dukla Pass. The possibility of the westward migration from Podole is also indicated (3). The grey area refers to the recent distribution range of the common hamster according to Mitchell-Jones et al. (1999). The distribution in Poland only was corrected according to the present changes in species range (Ziomek and Banaszek 2007)
The aim of this study is to extend the knowledge about the phylogeography of the common hamster and to explore colonization routes of central Europe by the species. The focus area is Poland because it is assumed to provide the dispersal corridor between the relatively strong German population and the eastern European core distribution area and naturally the historic expansion gate for the contemporary northern hamster lineage. Knowledge about genetic diversity of the common hamster in Poland is also valuable in the context of further conservation issues. Common hamster populations rapidly declined in many European countries (Nechay 2000; Neumann et al. 2005). In Poland, the studied species lost most of its former range in just 40 years and is now restricted to the south-eastern part of the country (Ziomek and Banaszek 2007). The common hamster is currently listed in Appendix IV of the Habitats Directive, which provides strict legal protection in all European Union countries. For further information see Data S1.
Material and Methods
Animals were collected from 12 populations in south-eastern Poland during the years 2005–2007 (Fig. 2, Table 1). Five populations are localized in the Lublin Upland (L1–L3) and Roztocze (R1–R2). This area is the biggest part of the contemporary, compact species range in Poland (Ziomek and Banaszek 2007). Two populations M1–M2 were sampled from the second largest area in the Malopolska Upland. The remaining five populations represent isolates at the western and southern limits of the current species range in Poland (Ziomek and Banaszek 2007): two populations from the Krakow–Czestochowa Upland (K1–K2), one site in Upper Silesia (U1) and two from the Sandomierz Basin (S1–S2); (Fig. 2, Table 1). The names of geographic regions follow Pucek and Raczyński (1983). The capture and handling of animals in the field was conducted under the permissions of the Minister of the Environment DOPog.-402-02-54/04/aj, Warsaw, Poland and the Local Ethics Committee on the Animal Research in Bialystok 2003/53, Bialystok, Poland.

The localization of sampling sites in Poland. (a) the contemporary common hamster range in Poland (Ziomek and Banaszek 2007). (b) the sampling sites belonging to Pannonia (P3) lineage are indicated by black circles, while open circles represent the E1 lineage. E1 site is from Neumann et al. (2005). The labels for sampling sites and geographic regions follow Table 1. The borders of the geographic regions are indicated with a dashed line
| Geographic region | Population | N | No. haplotypes | |||
|---|---|---|---|---|---|---|
| Comb. | ctr | 16S rRNA | cytb | |||
| Lublin Upland (LU) | L1 | 5 | 1 | 1CCdl361 | 116S122 | 1CbP9 |
| L2 | 6 | 3 | 1CCdl36 | 116S12 | 3CbP8, CbP9, CbP10 | |
| L3 | 5 | 3 | 1CCdl36 | 116S12 | 3CbP6, CbP7, CbP8 | |
| Roztocze (R) | R1 | 5 | 2 | 1CCdl36 | 116S12 | 2CbP8, CbP10 |
| R2 | 5 | 3 | 3CCdl36, Po1, Po3 | 216S12, 16SP1 | 4CbP5, CbP6, CbP8, CbP11 | |
| Malopolska Upland (MU) | M1 | 5 | 3 | 1CCdl36 | 116S12 | 3CbP8, CbP12,CbP13 |
| M2 | 6 | 1 | 1Po2 | 116S173 | 1Ccb104 | |
| Krakow–Czestochowa Upland (KCU) | K1 | 2 | 2 | 2Po2, Po4 | 116S17 | 1Ccb10 |
| K2 | 5 | 1 | 1Po2 | 116S17 | 1Ccb10 | |
| Upper Silesia (US) | U1 | 2 | 1 | 1Po2 | 116S17 | 1Ccb10 |
| Sandomierz Basin (SB) | S1 | 5 | 1 | 1Po5 | 116S17 | 1CbP4 |
| S2 | 4 | 3 | 2Po5, Po6 | 116S17 | 3CbP1, CbP2, CbP3 | |
| Poland | 55 | 17 | 7 | 3 | 14 | |
- 1Ccdl36 –AJ 633738, Neumann et al. (2005)
- 216S12 –AJ 633750, Neumann et al. (2005)
- 316S17 –AJ 633754, Neumann et al. (2005)
- 4Ccb10 –AJ 633765, Neumann et al. (2005)
We analyzed three partial mitochondrial (mt) regions: the control region (ctr), 16S ribosomal RNA (16S) and cytochrome b gene (cytb) in 55 animals. The ctr haplotypes were previously published (Banaszek et al. 2009). Total genomic DNA was extracted from frozen ear tips following a standard protocol supplied with the GenomicMini kit (A&A Biotechnology). PCR amplification followed the profiles described in Neumann et al. (2004, 2005). Amplified products were purified with CleanUp system microcolumns (A&A Biotechnology). Sequencing reactions were carried out using BigDyeTM Sequencing mix (v 3.1; Applied Biosystems, Foster City, CA, USA) in both directions. Amplification and sequencing reactions were performed in GeneAmp PCR System 9700 GOLD thermal cycler (Applied Biosystems). The sequencing reaction products were run on a 3130 Genetic Analyzer (Applied Biosystems).
In general, we used the primers chosen by Neumann et al. (2004, 2005). For cytb amplification and sequencing we had to use two different pairs of primers. Primers L14841 and HCRIC3 (Neumann et al. 2005) successfully amplified cytb in all individuals from the Lublin Upland, Roztocze and M1 population but most likely generated a nuclear pseudogene in other hamsters (populations: M2, K1-K2, U1, S1-S2), as the amplified sequence showed the anomalies commonly associated with the pseudogenes (Zhang and Hewitt 1996). Therefore, we used other primer pair: L14727-SP and H-ISO-SP (Jaarola and Searle 2002). For the phylogenetic trees and the parsimony network we used not only the sequences from Poland described in this paper, but also previously published sequences from the European species range (Neumann et al. 2004, 2005). The GenBank accesion numbers are the following: for ctr sequences –AJ550189, AJ550192, AJ550194, AJ550198, AJ550200–AJ550202, AJ550204, AJ550205, AJ633722, AJ633724–AJ633727, AJ633729–AJ633736, AJ633738, for cytb–AJ633756–AJ633782 and for 16S rRNA –AJ633739–AJ633754. All statistical and software information can be found in Data S1.
Results
In our previous study the ctr was sequenced for 195 individuals and only seven haplotypes (AJ 633738, EU 016106–EU 016111) were found (Banaszek et al. 2009). Considering the low variability of the ctr in our sample, we decided to use only 55 individuals for further analyses. The numbers of individuals examined for each population are given in Table 1. One ctr haplotype (AJ 633738) was previously reported by Neumann et al. (2005). Only one transversion was observed, the remaining seven substitutions were transitions in 366 bp sequence (Banaszek et al. 2009).
Fourteen cytb haplotypes were found in 55 hamsters. Thirteen were novel (EU107523–EU107535) and one was already reported (AJ 633765, Neumann et al. 2005). Twenty-three nucleotides were polymorphic among 906 bp of sequence. Twenty substitutions were transitions and only three transversions. Twenty variable sites were parsimony informative. The translation of the sequence resulted in a protein of 201 amino acids. Six out of 23 mutations were non-synonymous and led to amino-acid changes. Three mutations on the protein level were singletons and when plotted on the nucleotide sequence tree they occurred on the terminal branches, while the remaining three appeared on the internal tree branches and were characteristic for populations S1 and S2.
Three 16S haplotypes were identified in 55 animals. Two haplotypes were already described (AJ 633750, AJ 633754, Neumann et al. 2005) and one was new (EU107522). Four out of 468 nucleotides were variable and three of them were informative under parsimony. All substitutions were transitions. As a consequence of the relatively low genetic variability of our hamster populations we combined all three mt genes for further analyses to enhance statistical power. Altogether, we found 17 combined haplotypes in our Polish sample.
Network as well as phylogenetic tree consistently separate Polish hamsters into two lineages (3, 4). One lineage comprises samples L1–L3 from the Lublin Upland, R1–R2 from Roztocze and M1 from the northern part of the Malopolska Upland. The single hamster collected previously from the area of Poland, designed as the sample E1 (Neumann et al. 2005), belongs to this lineage (2, 3), therefore the whole group would be labelled E1. A second clade in Polish sample is formed by haplotypes from M2 sampling site (southern part of the Malopolska Upland), K1–K2 (Krakow–Czestochowa Upland), U1 (Upper Silesia) and S1-S2 (Sandomierz Basin) (Fig. 2 for geographic localization). There were no mixed populations with haplotypes characteristic for both lineages.
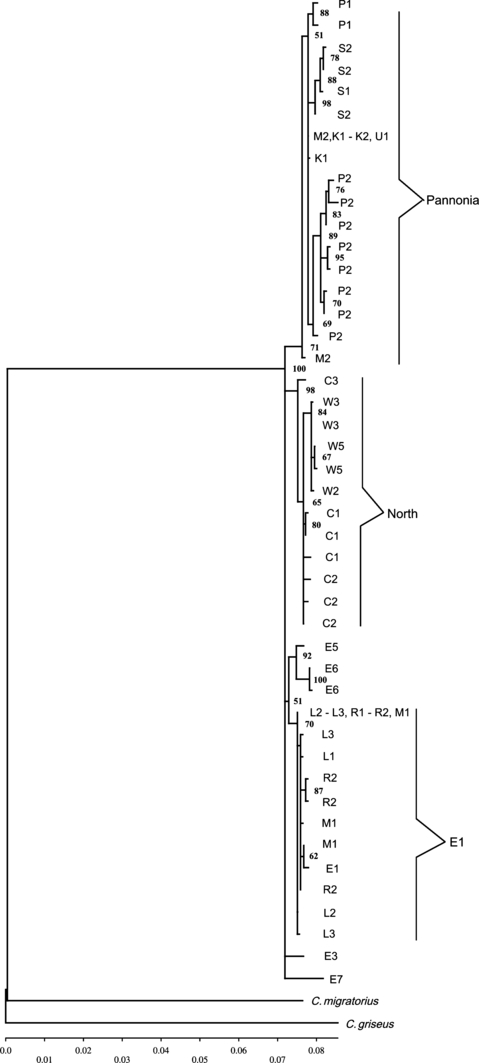
Neighbour-joining tree illustrating the phylogeographic relationships among the European common hamsters. Seventeen combined ctr + 16S + cytb haplotypes are from Poland and 27 from the rest of the species range (Neumann et al. 2005). Cricetulus griseus and Cricetulus migratorius served as outgroups. The bootstrap values (1000 replicates) are shown. The geographic location of the Polish haplotypes is indicated with population symbol as in Table 1. The geographic location of the European haplotypes follows the symbols on the Fig. 1, as in Neumann et al. (2005)
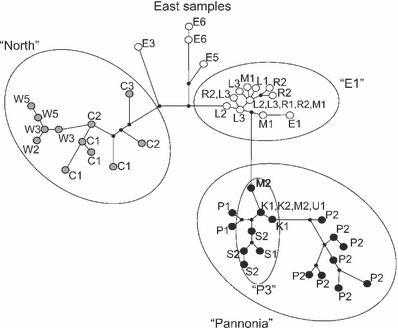
Parsimony network based on combined ctr + 16S + cytb haplotypes obtained from the European common hamsters. Seventeen haplotypes are from Poland and 27 from the rest of the species range (Neumann et al. 2005). Geographic location in case of Polish samples is indicated with the labels as in Table 1. The remaining haplotypes (Neumann et al. 2005) are labelled according to their geographic localization indicated in Fig. 1. The colour-coding for circles is consistent with 1, 2. The length of the links refers to the number of substitutions dividing haplotypes
The European hamsters are divided into two main clades: Pannonia lineage from the Carpathian basin and all the remaining populations comprising E1 hamsters from Poland, hamsters from Russia (other East samples) and specimens of the previously described lineage ‘North’ from central Europe (Fig. 3). The Polish E1 group clusters with Russian E5 and E6 samples. However the clade formed by E5 and E6 samples is characterized by very high bootstrap values, hence Polish E1 group and Russian samples E5 and E6 very probably form independent phylogeographic lineages. Due to scarcity of data from the eastern part of the species range, we cannot without doubt establish the relationships of Polish E1 group and other East samples. By contrast, second Polish lineage is nested within the Pannonian clade and following Neumann et al. (2005) nomenclature: P1 for Czech, P2 for Hungarian samples, it will be labelled P3 (3, 4).
The amova showed that genetic variation is mainly partitioned between two phylogroups (80.31%, p = 0.003), while variability between populations within group and within populations accounts only for 10.53% (p < 0.001) and 9.16% (p < 0.001) respectively. Net distance between E1 and P3 phylogroups based on the combined sequences was Da = 0.86 ± 0.21% (Table 2). The E1 and P3 lineages probably split ∼66–115 ka (95% CI: 34–170). Mean nucleotide divergence within phylogroups equals 0.09 ± 0.03% for E1 and 0.23 ± 0.03% for P3 and the difference suggests different demographic history of the groups.
| Sequence | Da % | E1 | P3 | All | |||
|---|---|---|---|---|---|---|---|
| N | π% | N | π% | N | π% | ||
| cytb | 1.23 ± 0.33 | 8 | 0.143 ± 0.102 | 7 | 0.447 ± 0.258 | 14 | 0.829 ± 0.436 |
| ctr | 0.58 ± 0.38 | 2 | 0.030 ± 0.055 | 5 | 0.203 ± 0.159 | 7 | 0.350 ± 0.245 |
| 16S | 0.65 ± 0.35 | 2 | 0.014 ± 0.033 | 1 | 0.000 | 3 | 0.326 ± 0.220 |
| Comb. | 0.86 ± 0.21 | 9 | 0.089 ± 0.061 | 8 | 0.312 ± 0.174 | 17 | 0.613 ± 0.314 |
The E1 lineage does not show any signs of further sub-structuring but a star-like network topology (Fig. 4). Pairwise mismatch analyses revealed a unimodal pattern for E1 (Fig. 5) indicating recent population expansion. Neutrality statistics is equivocal (all genes combined: Fs = −2.197, p = 0.093; cytb: Fs = −2.820, p = 0.037; 16S: Fs = −1.238, p = 0.05; ctr: Fs = 0.448, p = 0.667). However, it should be kept in mind that genetic variation is low in our sample and may therefore reduce the power of our statistical tests. Since we cannot exclude a scenario of recent expansion, we estimated the onset of population growth for E1 as 6.8–11.8 ka (95% CI: 1.6–18.4).

Pairwise mismatch distributions for E1 (a) and Pannonia (P3) (b, c) populations, which represent lineages with different glacial histories. The solid line describes the expected distribution of pairwise sequence comparisons that differed by a given number of nucleotides under the model of demographic expansion (a, b) and model of spatial expansion (c). The observed frequency is represented by vertical bars
On the other hand, the lineage P3 (Polish Pannonia) is clearly divided according to geographic regions (Fig. 2); populations from the southern part of the Malopolska Upland, Krakow–Czestochowa Upland plus Upper Silesia (M2, K1, K2, U1) form one group and two populations from the Sandomierz Basin (S1, S2) the other (3, 4). These two population groups share the same 16S haplotype but have non-overlapping ctr and cytb haplotypes (Table 1). Moreover, the populations from the Sandomierz Basin, S1 and S2, show four amino-acid changes within the cytb gene and are therefore differentiated from all other Polish haplotypes (Table 1).
The P3 lineage does not possess the genetic signature of recent demographic expansion (Fs = 1.359, p = 0.78; mismatch distribution goodness-of-fit test p = 0.050). The observed bimodal or ‘ragged’ distribution is clearly the result of the presence of two sublineages (Fig. 5). However, as they both without doubt belong to Pannonia lineage, we tested them together assuming a spatial expansion model and it gave a better fit of the expected pairwise mismatch distribution to the observations (test of goodness-of-fit, p = 0.36). A relatively recent range extension from a subdivided population appears to be a very likely scenario for the P3 group. The molecular dating for the spatial expansion of P3 is 29–50 ka (95%CI: 14–93 ka).
Discussion
The phylogeographic origin of Polish hamsters
In our analysis of the ctr variation in the group of populations defined now as E1 lineage we hypothesized that very low genetic variation was most probably caused by the purely accidental very small effective population size in the refugium for this phylogroup (Banaszek et al. 2009). However, more extensive analysis shows that it likely represents a western edge population of yet not identified populations of Ukrainian or Belarusian hamsters. Unfortunately, there is no paleontological record on rodent communities from the present E1 range in Poland, i.e. mostly the Lublin Upland, but climatic conditions throughout the Vistulian were very harsh, probably preventing hamsters to establish stable populations in that region. In this respect it appears very possible that the E1 group appeared very recently in Poland, according to our molecular dating about 6.8–11.8 ka (95% CI: 1.6–18.4). There is evidence for a rapid spread of steppe plant communities in eastern Poland e.g. during the Younger Dryas ∼12 ka (Madeyska 1995) providing favourable conditions for a range extension of the common hamster. Fossil data prove a continuous presence of hamsters in southern Ukraine (Crimea and Nikolayev province), from the Eemian interglacial and through the Vistulian (layers from Early and Late Vistulian) (Kowalski 2001). These populations may have served as sources of expansion, in particular because of the absence of significant geographical barriers.
The E1 phylogroup in Poland shows further signs of being the edge population. Nucleotide diversity of the E1 group is low and it shows star-like haplotype topology in network (Table 2, Fig. 4). Such characteristics make it similar to the ‘West’ common hamster sublineage of the North phylogroup, comprising the set of isolated populations on the western species border i.e. in the Netherlands, Belgium, France and Western Germany. The West common hamster sublineage most probably split from Central German populations during postglacial expansion (15–10 ka) (Neumann et al. 2005). Similar population structure indicates that both groups, West and E1, may have originated from leading edge dispersal events (Hewitt 1996). Lower diversity is generally expected for the populations from the border of the species range. However, the Polish populations of E1 lineage traditionally would be considered as situated far from the edges of the species range, while our molecular results showed that they had all the characteristics of the marginal populations. The expansion of the E1 group was further delayed by climate warming and forest development in eastern Poland during the Holocene (Madeyska 1995). The Neolithic which started not earlier than 6 ka in Poland (Wiślański 1970) provided the hamster with new suitable habitats because of the spread of agriculture and large scale wood clearances. Anthropogenic land use is therefore a key factor ensuring the survival of these marginal populations until now.
The Pannonia lineage in Poland is genetically more variable than the E1 lineage and mismatch as well as neutrality tests do not suggest demographic expansion (Fig. 5). However, there is evidence for a relatively recent spatial expansion from a sub-divided population according to the mismatch data (Excoffier 2004). In line with that is a haplotype frequency gradient from Hungary to Poland as expected for range extension (Barbujani et al. 1995; Ibrahim et al. 1996). The Pannonian lineage is located mainly in the Carpathian basin (Neumann et al. 2005). Fossil data and molecular clock estimates suggest the uninterrupted presence of the hamsters during the last 40–50 ka in the Hungarian plains (Janossy 1986; Kowalski 2001). Therefore, the Pannonian lineage in Poland is most likely the result of a northward range extension during the last glaciation. At this time the Carpathian Basin and the area of southern Poland remained ice-free (Lindner and Marks 1995). There are excellent fossil data about Pleistocene rodent communities from the caves and rock shelters situated in the southern part of the Krakow–Czestochowa Upland (Nadachowski 1989; Kowalski 2001). According to fossils, the common hamster did not occur in Poland during the first Vistulian stadial V1 (from 114.68 ka), as it disappeared at the beginning of the last glaciation i.e. Early Vistulian because of extreme cold and arid conditions. It reappeared in the Middle Vistulian V2 and during the stadials V2 (53.35 ka) and V3 (about 25 ka) occurred as an occasional and scarce element and was accompanied by steppe (sousliks, voles of arvalis type) and steppe-tundra elements (lemmings). Towards the end of the glaciation it was regularly documented for the Late Vistulian LV (about 15 ka) and it became even locally abundant in some assemblages (Nadachowski 1989; Kowalski 2001). Our molecular timing for the spatial expansion of the Polish Pannonia group, about 29–50 ka (95% CI: 14–93), roughly coincides with the paleontological timing of the common hamster fossils from the V2 and V3 stadials. Moreover, the timing for the spatial expansion of Pannonia into the area of Poland is also in agreement with the molecular dating of the demographic expansion of this group in the Carpathian basin, calculated by Neumann et al. (2005) for 25–44 ka (95% CI: 11–112). It indicates a very rapid colonization process and suggests the possibility to overcome strong geographical barriers by dispersers from the expanding populations under favourable climatic conditions.
Immigration from southern populations into Poland probably proceeded via two different routes, the Moravian Gate between the Carpathians and Sudetes, and the Dukla Pass directly through the Carpathians (Fig. 1). The routes have been used by a number of steppe plants of the Pontian-Pannonian type as well (Tacik 1959; Wróblewska 2008) and some relict steppe plant communities remained up today in the Malopolska and Krakow–Czestochowa Uplands (Pawłowski 2003). The Moravian Gate was most probably used by hamsters, which inhabited the Upper Silesia, Krakow–Czestochowa and the southern part of the Malopolska Uplands (1, 2). Five populations sampled from there were fixed for a single cytb haplotype (Table 1), which was also present in hamsters from Southern Moravia/Czech Republic (Neumann et al. 2005). The Polish side of the Moravian Gate, the area of the Glubczyce Plateau was densely populated by hamsters just 40 years ago but they became recently extinct (Surdacki 1969; Ziomek and Banaszek 2007). The second route, the Dukla Pass (500 m above see level), connects Slovakia with the Wislok and Wisloka river valleys on the Polish side, which similarly to Glubczyce plateau were inhabited by hamsters just 40 years ago (Surdacki 1970). Populations S1 and S2 are most probably the descendants of the hamsters which used that way of migration (1, 2). The only alternative colonization route would be through the Podole region (Fig. 1), although this would imply the presence of the Pannonian lineage in the southern Ukraine. Unfortunately, we have no genetic data from that region and further research is required to identify potential source populations in the more easterly distribution area of the common hamster.
The phylogeographic structure of the common hamster in Europe
Our study revealed that Polish hamsters comprise two distinct phylogeographic lineages, E1 and Pannonia (P3), with a clear north-south divergence pattern within the country (Fig. 2). Divergence time of the lineages E1 and P3 in Poland was calculated as 66–115 ka, which coincides fairly well with previous estimates of the North – Pannonia separation time (85–147 ka) and confirms the opinion that major European phylogroups already separated before they re-colonized Central Europe, most probably in the southern Russian and Ukrainian plains (Neumann et al. 2005). However, we still cannot resolve the colonization routes to the Western Europe, as no Polish haplotypes could be linked with German populations. As Neumann et al. (2005) suggested, the northern part of the species range could have been very unstable at the end of the last glaciation with very quick climatic oscillations during the Younger Dryas ∼12 ka. Several very rapid expansion and extinction events could wipe out the ‘North’ common hamsters from the area of Poland. On the other hand, the common hamster in Poland lost most of the range during the last 40 years and is now restricted to the south-eastern part of the country (Ziomek and Banaszek 2007). With such extensive loss of the species range, we cannot exclude the possibility that ‘North’ haplotypes were present in Poland in more northern located populations, which are extinct now.
The conservation issues
The common hamster is or becomes endangered in the scale of the whole continent (Nechay 2000). In Poland it is a strictly protected species under the Nature Conservation Act of April 16th, 2004 with an annotation that it demands active protection. The results obtained in this paper are important for the future management of natural hamster populations. The presence of two distinct lineages with different glacial histories should be kept in mind with any plans for reintroduction or creation of migration corridors, as mixing of such groups could be disadvantageous. The Polish Pannonian lineage is further subdivided and more research is needed to establish if the sublineages should be protected independently. Moreover, both Polish lineages have rather low genetic diversity caused by historical and contemporary bottlenecks (Banaszek et al. 2009). The remaining populations require quick conservation actions of creating hamster friendly agriculture areas (Nechay 2000) to prevent extinction.
Acknowledgements
The authors wish to express their gratitude to E Chwełatiuk, M Świsłocka, P Jadwiszczak and M Gąska, who helped in the field work. The work was financed by the Polish State Committee for Scientific Research under grant no. 2P04F 015 27 to A. Banaszek.




