Geographical patterns of mitochondrial DNA variation in Apis mellifera iberiensis (Hymenoptera: Apidae)
Patrones geográficos de variación del ADN mitocondrial en Apis mellifera iberiensis (Hymenoptera: Apidae)
Abstract
enAn extensive survey of mitochondrial haplotypes in honeybee colonies from the Iberian Peninsula has corroborated previous hypotheses about the existence of a joint clinal variation of African (A) and west European (M) evolutionary lineages. It has been found that the Iberian Peninsula is the European region with the highest haplotype diversity (12 haplotypes detected of the M lineage and 10 of the A lineage). The frequency of A haplotypes decreases in a SW-NE trend, while that of M haplotypes increases. These results are discussed in relation to hypotheses about the African origin of Apis mellifera and an early colonization of west Europe during intermediate Pleistocene glaciation events, followed by a regional differentiation. The extant pattern of haplotype frequency and distribution seems to be influenced at a regional scale by adaptation to local climatic conditions and the mobile beekeeping that has become a large-scale practice during the last decades. Other previous anthropogenic influences (Greek, Roman and Arab colonizations) are thought to be of minor importance in present day populations.
Resumen
esUn extenso estudio de los haplotipos mitocondriales en colonias de la abeja doméstica de la Péninsula Ibérica ha corroborado las hipótesis previas acerca de la existencia de una variación clinal conjunta de los linajes evolutivos africano (A) y europeo occidental (M). Se ha encontrado que la Peninsula Ibérica es la región europea con la mayor diversidad (12 haplotipos detectados pertenecientes al linaje M y 10 al linaje A). La frecuencia de los haplotipos africanos disminuye en la orientación SW-NE, al tiempo que aumenta proporcionalmente la de los M. Estos resultados se analizan en relación a las hipótesis recientes que ubican el origen de Apis mellifera en África, junto con otras que postulan una colonización temprana de esta especie en Europa occidental, seguida de una diferenciación durante el Pleistoceno. El patrón geográfico actual de haplotipos y frecuencias a escala regional, parece estar influido por la adaptación a las condiciones climáticas locales y la trashumancia, práctica que ha adquirido grandes proporciones en las últimas décadas. Otras influencias antrópicas acontecidas como las colonizaciones de griegos, romanos y árabes han tenido posiblemente poca influencia sobre las poblaciones ibéricas actuales.
Introduction
The honeybee Apis mellifera L. shows a remarkable regional differentiation, as 29 subspecies are known from Africa and the western Palearctic region (Engel 1999; Sheppard and Meixner 2003). Five evolutionary lineages have been characterized based on morphometric, molecular, ecological, ethological and physiological traits (review in De la Rúa et al. 2005b). Four of those occur naturally in the Mediterranean Basin: African lineage (A), West and North European lineage (M), South-east Europe lineage (C) and Near and Middle East lineage (O). The honeybees of the Iberian Peninsula are currently considered to be a particular subspecies, A. m. iberiensisEngel, 1999, originated after a natural hybridization between lineages A and M. Since the first morphometric and behavioural studies (summarized by Ruttner 1988) a number of authors studying enzymes (Smith and Glenn 1995; Arias and Sheppard 2005), pheromones (Hepburn and Radloff 1996), and mitochondrial DNA and microsatellites (Smith et al. 1991; Arias and Sheppard 1996; Franck et al. 1998; Garnery et al. 1991, 1998, 1998a,b; De la Rúa et al. 1999, 2002b, 2004a,b, 2005a; Cánovas et al. 2002; Arias et al. 2006) have supported the hypothesis that Iberian honeybees are the result of a wide intergradation between honeybees of the M branch that survived the last glaciation event, and North African honeybees of the A lineage that have colonized south-west Europe. In this hypothesis it is assumed that bees of the M lineage survived the last glaciation in different refugia of Iberia and started a northwards colonizing expansion in the last postglacial era, that in the same period there have been one or more colonization waves of honeybees coming from Africa (A lineage), and that hybridization between both lineages has given rise to a clinal distribution of haplotypes A and M, that is more gradual to the east of Iberia and sharp to the northwest (De la Rúa et al. 2005a,b).
In a recent study based on the analysis of single nucleotide polymorphisms Whitfield et al. (2006) have convincingly argued that Apis mellifera‘originated in Africa and that there were at least two subsequent expansions into Eurasia: a western expansion into Europe (lineage M) and one or more (independent) eastern expansions into Asia and east Europe (lineages O and C)’. This hypothesis demands a re-evaluation of previous ideas about the history of A. mellifera in the Iberian Peninsula, that should be traced to Pleistocene glaciation events, and that has been possibly influenced in recent historical times by beekeeping.
Garnery et al. (1998a) and Franck et al. (1998) suggested that Greek and Moorish colonizations of the Iberian Peninsula may have influenced present haplotype distribution. However, De la Rúa et al. (2002b, 2004b) postulated that these influences are negligible and that the finding of an mtDNA gradient across the whole Iberian Peninsula should corroborate its natural origin, as a man-made origin can hardly explain this geographic pattern on such large scale. To test this hypothesis we have made an extensive survey of the mtDNA of Iberian populations, paying particular attention to populations located in geographic gaps not yet investigated to date Further, a detailed analysis of the geographic distribution of African sublineages, AI, AII, and AIII (as defined by Franck et al. 2001) in the Iberian Peninsula is needed to test hypotheses about colonization waves. Finally, a comprehensive haplotype distribution map for the Iberian Peninsula may serve as an accurate basis for further evolutionary studies on a regional scale, e.g. the influence of selective factors on haplotype distribution.
Materials and Methods
Sampling and DNA extraction
Samples of 1017 colonies from 109 localities grouped according to Spanish provinces (Table 1 and Fig. 1) were studied. Adult workers from the inner part of the hives were sampled to avoid collecting drifting honeybees. Honeybees were transported to the laboratory in ethanol. Total DNA was extracted from one leg following Walsh et al. (1991) with a 5% Chelex-based protocol.
| Province | N | A1 | A2 | A3 | A4 | A6 | A8 | A9 | A11 | A14 | A15 | M2 | M3 | M4 | M4P | M5 | M6 | M7 | M7P | M8 | M8P | M9 | M12 | AI | AII | AIII | A | M |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Almería (6) | 89 | 9 | 68 | 0 | 0 | 0 | 3 | 0 | 0 | 2 | 0 | 0 | 0 | 6 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 77 | 3 | 2 | 82 | 7 |
| Asturias (13) | 109 | 4 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 65 | 0 | 0 | 8 | 5 | 0 | 14 | 0 | 5 | 4 | 8 | 0 | 0 | 8 | 101 |
| Ávila (2) | 19 | 0 | 1 | 3 | 0 | 0 | 8 | 4 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 4 | 12 | 1 | 17 | 2 |
| Burgos (10) | 48 | 1 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 0 | 16 | 11 | 8 | 0 | 3 | 0 | 0 | 0 | 4 | 0 | 0 | 4 | 44 |
| Cáceres (1) | 8 | 0 | 0 | 4 | 0 | 0 | 1 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 4 | 0 | 8 | 0 |
| Cádiz (12) | 70 | 1 | 59 | 0 | 3 | 1 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 64 | 0 | 5 | 69 | 1 |
| Ciudad Real (2) | 9 | 0 | 6 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 1 | 0 | 7 | 2 |
| Córdoba (6) | 87 | 16 | 54 | 2 | 0 | 0 | 5 | 3 | 0 | 4 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 72 | 8 | 6 | 86 | 1 |
| Granada (7) | 101 | 1 | 73 | 7 | 0 | 0 | 0 | 1 | 0 | 17 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 81 | 1 | 17 | 99 | 2 |
| Guadalajara (4) | 40 | 0 | 9 | 6 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 14 | 0 | 5 | 1 | 0 | 0 | 2 | 0 | 0 | 0 | 16 | 2 | 0 | 18 | 22 |
| Huelva (2) | 27 | 3 | 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 14 | 0 | 0 | 0 | 0 | 0 | 12 | 0 | 0 | 12 | 15 |
| Huesca (6) | 43 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 37 | 1 | 0 | 0 | 3 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 43 |
| Jaén (2) 60 | 5 | 39 | 1 | 0 | 0 | 6 | 4 | 0 | 1 | 0 | 1 | 0 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 45 | 10 | 1 | 56 | 4 | |
| Madrid (1) | 10 | 0 | 7 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 8 | 1 | 0 | 9 | 1 |
| Málaga (4) | 48 | 1 | 20 | 13 | 3 | 0 | 0 | 5 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 37 | 5 | 5 | 47 | 1 |
| Salamanca (2) | 19 | 2 | 10 | 3 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 15 | 2 | 0 | 17 | 2 |
| Santander (5) | 39 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 21 | 0 | 0 | 7 | 0 | 0 | 0 | 0 | 4 | 1 | 0 | 0 | 1 | 38 |
| Sevilla (6) | 51 | 6 | 27 | 0 | 0 | 1 | 8 | 5 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 34 | 13 | 2 | 49 | 2 |
| Teruel (6) | 47 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 32 | 3 | 0 | 0 | 8 | 0 | 0 | 0 | 0 | 1 | 3 | 0 | 0 | 3 | 44 |
| Zaragoza (12) | 93 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 47 | 3 | 0 | 0 | 30 | 5 | 1 | 2 | 0 | 5 | 0 | 0 | 0 | 0 | 93 |
| Total | 1017 | 50 | 392 | 40 | 7 | 2 | 36 | 26 | 1 | 36 | 2 | 1 | 1 | 220 | 30 | 22 | 21 | 81 | 5 | 23 | 2 | 5 | 14 | 491 | 62 | 39 | 592 | 425 |
- Values in brackets show the number of apiaries sampled in each province. N is the number of colonies analysed from each province.
Geographical origin of the honeybee populations sampled in the present work
PCR amplification and digestion
The mitochondrial DNA fragment including the tRNAleu-COII intergenic region was amplified according to a protocol detailed in Garnery et al. (1993). Two μl of the PCR product were run on 1% agarose gel for size determination and the remaining volume was incubated with 5 units of the restriction enzyme DraI at 37°C for 2–8 h. Restricted DNA fragments were discriminated on 3% agarose NUSIEVE® gels and stained with ethidium bromide.
Statistical analysis
Previous data obtained by French colleagues (Garnery et al. 1998a; Franck et al. 2001) on populations located in the Basque Country, Central Spain (Segovia, Toledo), Andalusia (Seville) and Porto (Portugal), and those formerly investigated in our laboratory (De la Rúa et al. 1999, 2001a, 2004b, 2005a; Cánovas et al. 2002) from Murcia, Balearic Islands, Valencia, Catalonia and Galicia, have also been added. Detailed data on these samples are available on request.
Unbiased estimates and standard deviations of gene diversity of mitochondrial DNA (D) were calculated following Nei (1987). To investigate relationships between populations and samples, minimum spanning networks were performed with the arlequin software (Schneider et al. 2000) using a square distance matrix. Alternative connections (see below) between haplotypes were explored.
A neighbour-joining tree based on the DA distance was calculated to establish optimally phylogenetic relationships among populations with different origins, by using the software Populations v. 1.2.29 (Langella 2005).
Results
Twenty-two haplotypes have been found after the RFLP analysis of the tRNAleu-COII intergenic region, 10 of them were from lineage A and 12 from lineage M (Table 1). The haplotypes were identified following previous reports (Garnery et al. 1993; Franck et al. 1998; De la Rúa et al. 2001a, 2004b, 2005a). Fifty-eight per cent of the colonies studied belong to the African lineage. Sublineage AI (haplotypes A1, A2, A3, A4 and A6) is the most common (83% of total lineage A) and widespread, followed by sublineage AIII (8.7%, haplotypes A11, A14 and A15) and AII (8.3%, haplotypes A8 and A9). Sublineages AI and, to a lesser extent, AII are widely distributed across the Iberian Peninsula (Fig. 3), whereas sublineage AIII is frequent in the south (Andalusia) and on the Atlantic coast of the Iberian Peninsula. These A lineages decrease in frequency across the whole peninsula following a SW-NE clinal variation.
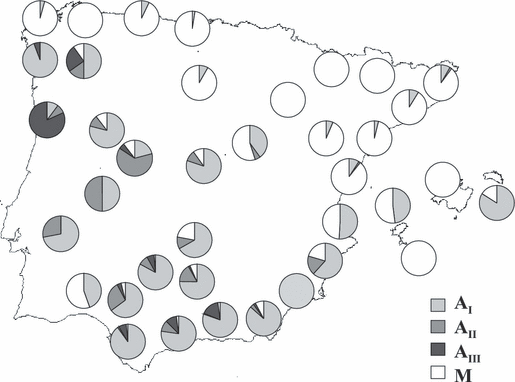
Map showing the distribution of mitochondrial evolutionary lineages and sublineages in the Iberian Peninsula. Data from Garnery et al. (1998a), De la Rúa et al. (1999, 2001a, 2004b, 2005a), Franck et al. (2001) and Cánovas et al. (2002) have also been included
A2 is the most frequent African haplotype (31%) and is present in all Iberian areas where the African lineage is found except Cáceres, Ciudad Real and Santander. M4 is the most frequent M haplotype and is distributed in all the northern Spanish provinces. Its frequency decreases southwards but is still present in five of the eight Andalusian provinces (south Iberia). A mixture of both A and M haplotypes occurs in almost all provinces except Zaragoza and Huesca (only M) and Cáceres (only A). Some haplotypes seem to be restricted to particular provinces: A11 (Ávila), M3 (Ciudad Real) and M2 (Jaén). A number of A haplotypes present in the Iberian Peninsula have not yet been reported from continental African populations, i.e. A28 in Alicante and Gerona (De la Rúa et al. 2004b, 2005a), A14 in all Andalusian populations except Huelva, and A15 in Cordoba. The last two haplotypes were initially described as typical of Canarian populations (De la Rúa et al. 2001b, 2002a).
The minimum spanning networks (Excoffier et al. 1992) for intrapopulation haplotype variation are shown in Fig. 2. The two M and A mitochondrial lineage networks are disconnected because of the high divergence between them. For the A lineage the minimum spanning tree was extended into a minimum spanning network (Fig. 2a), in which the alternative connections than link haplotypes A8 with A9 (sublineage AII) and A14 and A15 (sublineage AIII) are represented, in congruence with the sublineage division suggested by Franck et al. (2001). The haplotype A2 is located in a central position in the A lineage network. Other haplotypes from the same sublineage AI (A1, A3 and A4) are closely related though show varying frequencies in Iberian populations. In the minimum spanning network for the M lineage (Fig. 2b), the haplotype M4 is found in a central position and M7 is closely related to it. The Iberian populations show high haplotype diversity (D), with an average of 0.577; only four provinces have a D value lower than 0.400 (Cadiz, Huesca, Teruel and Zaragoza) (Table 2). This diversity remains high even when the two lineages are considered separately.
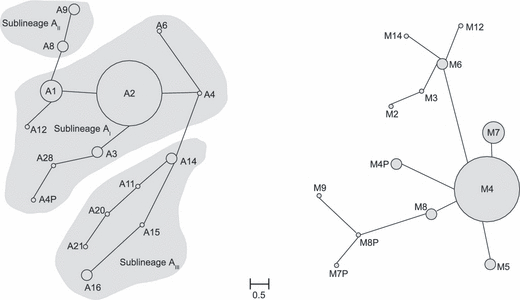
Minimum spanning network of the African (A) and west European (B) haplotypes found in the Iberian Peninsula. Frequencies of haplotypes are proportional to the circle radius. Links between haplotypes are proportional to genetic distances between them
| Provinces | Total | Lineage A | Lineage M | ||||||
|---|---|---|---|---|---|---|---|---|---|
| n | D | σf 2D | n | D | σf 2D | n | D | σf 2D | |
| Almería | 89 | 0.403 | 0.062 | 82 | 0.302 | 0.062 | 7 | 0.000 | 0.000 |
| Asturias | 109 | 0.565 | 0.052 | 8 | 0.571 | 0.094 | 101 | 0.495 | 0.055 |
| Ávila | 19 | 1.000 | 0.072 | 17 | 0.728 | 0.083 | 2 | ||
| Burgos | 48 | 0.758 | 0.029 | 4 | 44 | 0.716 | 0.027 | ||
| Cáceres | 8 | 0.679 | 0.122 | 8 | 0.679 | 0.122 | |||
| Cádiz | 70 | 0.286 | 0.069 | 69 | 0.265 | 0.068 | 1 | ||
| Ciudad Real | 9 | 0.583 | 0.183 | 7 | 0.286 | 0.196 | 2 | ||
| Córdoba | 87 | 0.580 | 0.054 | 86 | 0.570 | 0.055 | 1 | ||
| Granada | 101 | 0.448 | 0.054 | 99 | 0.426 | 0.054 | 2 | ||
| Guadalajara | 40 | 0.803 | 0.038 | 18 | 0.660 | 0.078 | 22 | 0.558 | 0.101 |
| Huelva | 27 | 0.590 | 0.063 | 12 | 0.409 | 0.133 | 15 | 0.000 | 0.000 |
| Huesca | 43 | 0.135 | 0.069 | 43 | 0.135 | 0.069 | |||
| Jaén | 60 | 0.563 | 0.072 | 56 | 0.499 | 0.076 | 4 | ||
| Madrid | 10 | 0.533 | 0.180 | 9 | 0.417 | 0.191 | 1 | ||
| Málaga | 48 | 0.742 | 0.043 | 47 | 0.731 | 0.043 | 1 | ||
| Salamanca | 19 | 1.000 | 0.010 | 17 | 0.632 | 0.112 | 2 | ||
| Santander | 39 | 0.603 | 0.053 | 1 | 38 | 0.582 | 0.051 | ||
| Sevilla | 51 | 0.682 | 0.060 | 49 | 0.656 | 0.063 | 2 | ||
| Teruel | 47 | 0.273 | 0.083 | 3 | 44 | 0.172 | 0.074 | ||
| Zaragoza | 93 | 0.307 | 0.060 | 93 | 0.307 | 0.060 | |||
A neighbour-joining tree computing all Iberian samples shows three main peninsular groups and a fourth group comprised of insular populations (Ibiza and Formentera) (Fig. 4). Group 1 corresponds to the southern populations (Andalusia) together with others from central Iberia (Salamanca, Ciudad Real, Madrid). Group 2 includes the populations of the two southern Balearic Islands (Ibiza, Formentera) and is related to group 1. Group 3 is made up by populations from north-western Spanish provinces together with Portuguese samples (Porto and Évora) and some from central Spain (Cáceres, Ávila,). Honeybee populations from Majorca and Minorca show DA values related to this last group. To the north there is a fourth group made up by populations of north Iberia (Huesca, Lérida, Teruel, Castellón, Gerona, Asturias, Barcelona, Zaragoza, Tarragona, and Santander) and Guadalajara (central Spain).
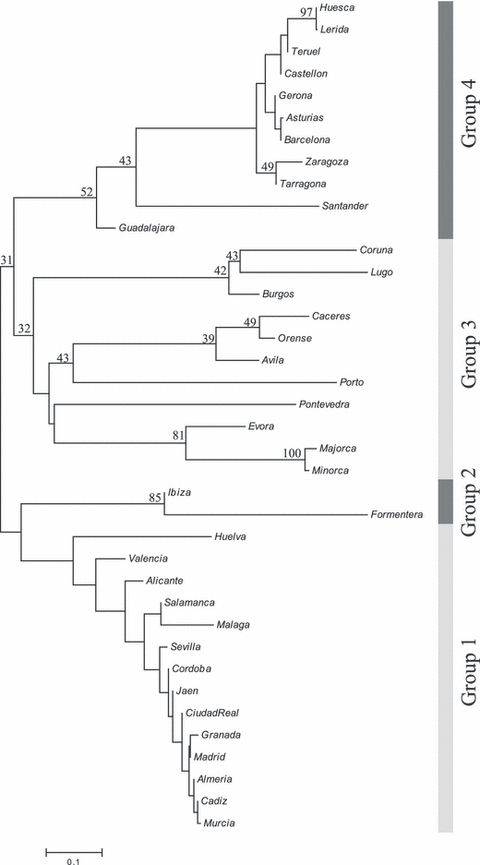
Unrooted Neighbour-joining tree of honeybee Iberian populations based on the distance DA for mitochondrial data. Bootstrap values have been computed over 2000 iterations by resampling individuals within samples and are noted as percentages. Data reported by Franck et al. (1998, 2001), De la Rúa et al. (1999, 2004b, 2005a) and Cánovas et al. (2002) have also been included
Discussion
More than 2000 colonies of Apis mellifera iberiensis have been studied to date, revealing the occurrence of 12 haplotypes of the M lineage and 10 of the A lineage. Thus the Iberian Peninsula is the region of Europe with the highest diversity of mitochondrial haplotypes known so far. As a comparison Franck et al. (1998) reported 11 haplotypes in a transect from the Pyrenees to the Scandinavian Peninsula. Likewise, these authors reported 23 A haplotypes in the whole African continent, where 11 subspecies are known. African haplotypes found in Iberia correspond to the three sublineages AI, AII and AIII established by Franck et al. (2001). This high diversity shows a definite geographic pattern as it has been corroborated that there is a clinal replacement of A haplotypes by M haplotypes following a SW-NE transect across the Iberian Peninsula. This transect was first suggested by Garnery et al. (1998a) based on a limited number of samples.
The transition from A to M haplotypes occurs in the whole Iberian Peninsula, thus supporting the hypothesis of a natural origin of the pattern at a peninsular scale. According to Whitfield et al. (2006) there was a particular colonization wave of west Europe by honeybees coming from North Africa. An approximate estimation of Garnery et al. (1992) about the time of divergence between A and M lineages is of 1.25 Myr, which traces back the origin of the M lineage to the Günz glaciation (1.1 Myr). According to Arias and Sheppard (1996) the estimate for this event ranges from 0.85 to 0.47 Myr. Both calculations allow for the first A colonizers to differentiate into the M lineage after successive episodes of extinction, survival in refugia and re-colonization of west Europe that occurred during intermediate Pleistocene climatic oscillations. This hypothesis is congruent with the fact that the subspecies A. m. mellifera show nowadays the ability of colonizing temperate and cold regions of Europe (Garnery et al. 1998a) by means of morphological and ethological adaptations (Ruttner 1988). A marked decrease of haplotype diversity in the M lineage from North Spain to Scandinavia and the microsatellite data indicate the occurrence of a population bottleneck that also supports this hypothesis (Franck et al. 1998; Garnery et al. 1998a,b, present paper). The M4 haplotype is perhaps ancestral because of its widespread distribution and high frequency in Iberian and south French populations. Other A. m. mellifera populations probably survived in Italy but are not good candidates for ancestors of European populations, because they only show the M7 haplotype (Franck et al. 2000) that is rare elsewhere.
The first colonizers possibly belong to the AI sublineage, as inferred from its high frequency and widespread distribution in Iberia and the Mediterranean Basin. Populations of this sublineage have been reported in Balearic Islands (De la Rúa et al. 2001a), Sicily and some Greek Islands (Garnery et al. 1993). It is interesting to note that the predominant A2 haplotype of this sublineage is nowadays rare in North Africa (Garnery et al. 1995; Barour et al. 2006; De la Rúa et al. 2007). Honeybees belonging to the sublineage AII (A8–A10) possibly came later as they are less frequent and their geographical distribution is not so widespread. The last sublineage to arrive was possibly AIII (A14–A16), because it is restricted to the western parts of the Peninsula. This last lineage shows an Ibero-Atlantic pattern of distribution already described for other insect groups (Serrano et al. 2003), and is also found in the Macaronesian archipelagos (De la Rúa et al. 2001b, 2002a, 2006) and the Moroccan Atlantic coast (De la Rúa et al. 2007).
Additional factors must be invoked to explain the haplotype distribution found in our survey at a regional scale (2, 3, 4). Preliminary results indicate that climatic factors are exerting a selective pressure on the whole genome of A and M honeybees (F. Cánovas, P. De la Rúa, J. Serrano, J. Galián, unpublished data). Another factor that is certainly influencing the haplotype distribution is the practice of transhumance and the increased turnover of colonies caused by the varroasis during recent decades. Transhumance affects today about 80% of the more than 2.4 million of Spanish colonies. Inadequate and extreme management causes many losses that lead to colony replacement by purchasing queens, often from other regions (P. De la Rúa, J. Serrano, unpublished data). This turnover has been reinforced by the need of recovering hives decimated by varroasis (which spreading has been in turn favoured by transhumance). Although the mitochondrial haplotype has probably been locally stable for centuries because of its maternal inheritance, the man-made replacement of native colonies may rapidly change the local haplotype composition, as we found in the islands of Formentera and Ibiza (De la Rúa et al. 2001a), or in the Canary Islands (De la Rúa et al. 2002a). This practice is also the most likely explanation for the finding of M haplotypes in provinces of South Spain with mild climates (Almería, Huelva, etc.), where the beekeepers have confirmed the purchase of colonies from North Spain. Therefore, whether both historical and climatic factors would produce a natural distribution pattern of haplotypes across the Iberian Peninsula, anthropogenic factors may rapidly change this pattern because of the great development of mobile beekeeping in the last two decades. The moderate geographic incongruence observed in the unrooted neighbour-joining tree of Fig. 4 is the likely result of these recent changes, and the prediction is that the pattern may be substantially altered in a few years.
Finally, Garnery et al. (1995, 1998a) and Franck et al. (1998) have suggested that the introduction of colonies from North Africa during recent historical times by Ancient Greeks, Romans and Arabians may have influenced the pattern found for A haplotypes in Iberia. Likewise, Sheppard et al. (1996) indicated that ‘the potential role of human-assisted movement of African A. m. intermissa/sahariensis into Iberia during 800 years of Moorish occupation cannot be dismissed without careful historical investigation’. However, the frequency and diversity of A haplotypes clearly differs from Morocco and Algeria to South Spain (Garnery et al. 1995; Arias and Sheppard 1996; De la Rúa et al. 2004a, 2007; Barour et al. 2006; present paper), and suggests that the effective gene flow between both areas has been low. This conclusion is supported by both selective and historical factors. First, the effect of local conditions on the fate of imported colonies was noted by Garnery et al. (1993, 1995), who reported the absence of M haplotypes in Algeria in spite of repeated importations from France for decades. With regard to importations during the Arab colonization of the Iberian peninsula (VIII–XV centuries), it should be noted that the most important human movements from North Africa to Iberia were the entrance of Arab armies in the VIII, XII and XIII centuries, who had, presumably, little apicultural significance. In fact, the human movement that could have a remarkable apicultural effect was the massive expulsion of Moorish people from Spain to North Africa during the XVI and XVII centuries. About 300 000 people were forced to abandon different Spanish regions and moved to North Africa during 1570 and 1609–1614 (Lea 1968), with the possibility of bringing some baggage, including hives. However, no markers of these hypothetical tradeoffs have been found, perhaps because of drift and selection, thus meaning a historical event of scarce importance for present-day populations.
Acknowledgements
Thanks are due to the many beekeepers and associations who generously collaborated with us in collecting the samples. Two reviewers much improved the ms with valuable comments. Specially thanks to O. Rodríguez, J. Ornia, U. Mediel and COAG association. Thanks to Dr O. Langella for his support with Populations software. This research has been supported by the following projects: RZ00-013 (INIA, Instituto Nacional de Investigaciones Agrarias, INIA, Spain), EVK2 2000-00628 (European 5th Framework), BOS2003-9765 (Ministerio de Educación y Ciencia, Spain) and API06-010 COORD-2 (Ministerio de Agricultura, Pesca y Alimentación, Spain).




