Species in the genus Turritopsis (Cnidaria, Hydrozoa): a molecular evaluation
Indagine molecolare delle specie del genere Turritopsis (Cnidaria, Hydrozoa)
Abstract
enMitochondrial ribosomal gene sequences were used to investigate the status of several populations of hydromedusae belonging to the genus Turritopsis (family Oceaniidae). Several nominal species have been described for this genus, but most of them had been synonymized and attributed to one cosmopolitan species, Turritopsis nutricula. A recent revision based on morphological and reproductive characters, however, has shown that many different populations can be distinguished and that several of the nominal Turritopsis species are likely valid biological species. Our investigation using molecular sequence data of 16S mitochondrial gene confirms these results. The Mediterranean Turritopsis must be attributed to Turritopsis dohrnii and the Turritopsis of New Zealand must be referred to Turritopsis rubra. The situation of the Japanese Turritopsis is more complex, though all sampled populations are clearly distinct from T. nutricula, a species likely confined to the Western Atlantic. The Japanese Turritopsis fall into three widely separated lineages. One of them, corresponding likely to Turritopsis pacifica, is closely related to T. rubra. A second clade, which potentially represents an as yet undescribed species, produces smaller medusae than T. pacifica and is morphologically distinguishable from it. Finally, a third group was distinguished by a single haplotype sequence that is identical with a Mediterranean sample of T. dohrnii. It is postulated that the last group of Japanese Turritopsis is likely a recent introduction, most probably by human activity. A survey of all known and potentially valid Turritopsis species is given in table format to facilitate identifications and future revisory work.
Sommario
itSequenze del gene mitocondriale 16S sono state utilizzate per studiare lo stato tassonomico di idroidi appartenenti al genere Turritopsis (Famiglia Oceaniidae). In letteratura, tra le numerose specie nominali di TurritoSPSis descritte, molte di queste sono state successivamente messe in sinonimia e attribuite ad un'unica specie cosmopolita, Turritopsis nutricula. Una recente revisione, basata su dati morfologici e caratteri riproduttivi, ha comunque mostrato che diverse popolazioni di Turritopsis possono essere distinte in numerose specie nominali e probabilmente rappresentano valide specie biologiche. Il presente studio conferma questa recente interpretazione, mediante lo studio di sequenze molecolari del gene 16S. La popolazione mediterranea di Turritopsisè ora attribuita a T. dohrnii, mentre la popolazione neozelandese va ascritta alla specie T. rubra. La situazione nei mari giapponesi si presenta piu’ complessa, sebbene tutte le popolazioni ivi campionate siano chiaramente distinte da T. nutricula, la quale risulta confinata unicamente all'Atlantico Orientale. Le sequenze ottenute da esemplari di Turritopsis provenienti dal Giappone formano tre cladi ben distinti. Uno di essi corrisponde a Turritopsis pacifica. Un secondo clade è costituito da popolazioni che producono meduse piu’ piccole rispetto a Turritopsis pacifica ed e’ dunque anche morfologicamente separato. Un terzo gruppo e’ rappresentato da un solo aplotipo identico alle popolazioni mediterranee di T. dohrnii. La presenza di quest'ultimo gruppo di Turritopsis in Giappone e’ molto probabilmente il risultato di un'introduzione recente, in seguito ad attività umana. Per facilitare futuri lavori di revisione, è inoltre presentata tavola che riassume le caratteristiche di tutte le specie di Turritopsis conosciute e potenzialmente valide. La tavola cerca di integrare i dati morfologici e riproduttivi già noti e dei dati molecolari ottenuti con questo studio.
Introduction
Medusae of the genus Turritopsis occur all around the globe in temperate to tropical waters. In some localities, e.g. the English Channel and New Zealand, they can be quite conspicuous due to their red colour and well-known to beach-goers, fishermen and naturalists. In recent years, Turritopsis has also found his way into the ordinary daily press and scientific journals as it was portrayed as being potentially immortal due to its ability to revert its life cycle (Bavestrello et al. 1992; Piraino et al. 1996, 2004). One might expect that the taxonomic status for such an animal is clear and well-settled, as this is a crucial prerequisite to know to what extent the results of these experimental studies can be generalized. However, as shown by Schuchert (2004) and the present study, the species boundaries within the genus Turritopsis are by no means settled and its taxonomy still needs to be clarified by further investigation.
Several nominal species referable to the genus Turritopsis have been described (see the taxonomic section), but recent authors followed the authorities of Mayer (1910), Kramp (1961) and Russell (1953) in regarding only a few species as valid. Kramp (1961) kept only two nominal species and synonymized most of the names with Turritopsis nutricula. Turritopsis nutricula was hence regarded as a species with a circumglobal distribution. Slight differences among the various populations have been noted and acknowledged (e.g. Russell 1953), but they were considered as representing intraspecific variation. Recently, Schuchert (2004) showed that Turritopsis populations from the North-Eastern Atlantic, the Mediterranean, and New Zealand have distinct morphologies and life histories from those of the American T. nutricula. They were therefore assigned to four separate species.
The case of Turritopsis also illustrates a general problem in hydrozoan systematics: nominal species are mostly based either on the polyp or on the Medusa phase alone, thus with insufficient knowledge of the entire life cycle. The first complete life cycle described for a species of Turritopsis was the one of T. nutricula (see Brooks 1886). Subsequent authors then often identified similar hydroid colonies from various other regions of the earth as belonging to T. nutricula, generally without knowing the complete life cycle. However, almost identical hydroids may produce very different medusae, sometimes even belonging to different families (e.g. Pandeidae and Bougainvillidae, Corynidae and Cladonematidae, hydroids of the Campanulina type).
With the advent of molecular genetic studies it has become clear that cryptic speciation and sibling species are rife in the marine realm (Knowlton 1993). For the Hydrozoa, this has been shown in very few cases, the most prominent being the discovery of sibling species in the genera Hydractinia (Buss and Yund 1989), Obelia (Govindarajan et al. 2005a), Eugymnanthea (Govindarajan et al. 2005b) and Coryne (Schuchert 2005). As there is no reason to believe that other hydrozoan genera should not parallel the cryptic speciation already found, the species diversity within Turritopsis can be expected to be larger than previously thought.
The aim of this investigation was therefore to evaluate the various Turritopsis populations and species by molecular methods and to test some of the conclusions reached by the morphological examinations of Schuchert (2004). A part of the sequence (615 bp) of the mitochondrial RNA gene (16S) was used because it has been demonstrated to be useful to identify hydrozoan species (Cunningham and Buss 1993; Govindarajan et al. 2005a,b). Additionally, all potentially valid species of the genus Turritopsis are reviewed to facilitate the discussion and future revisory work.
Materials and Methods
Data for all samples used in this study are given in Table 1. Voucher specimens for the Mediterranean and New Zealand animals were deposited in the Natural History Museum of Geneva, Switzerland (MNHG).
| Sequence name | Species identification | Locality, | Date | Material | MHNG voucher | EMBL accession number |
|---|---|---|---|---|---|---|
| Andalucia | Turritopsis spec. | Mediterranean, Spain, Andalucia, Las Negras | 28 July 2003 | Monosiphonic colony | – | – |
| Mallorca 1 | Turritopsis dohrnii | Mediterranean, Mallorca, Cala Murada | 15 July 1997 | Polyps | – | 3 |
| Mallorca 2 | Turritopsis dohrnii | Mediterranean, Mallorca, Cala Murada | 16 August 2000 | Polyps | 29753 | AY787889 |
| Mallorca 3 | Turritopsis dohrnii | Mediterranean, Mallorca, Cala Murada | 22 August 1999 | Polyps | 27123 | – |
| New Zealand 1 | Turritopsis rubra | New Zealand, Wellington Harbour | 12 July 2002 | Polyps | – | – |
| New Zealand 2 | Turritopsis rubra | New Zealand, Hauraki Gulf | 29 July 2002 | Medusae | 33469 | Identical to T. NZ1 |
| New Zealand 3 | Turritopsis rubra | New Zealand, Hauraki Gulf | 29 July 2002 | Medusae | 33469 | Identical to T. NZ1 |
| New Zealand 4 | Turritopsis rubra | New Zealand, Hauraki Gulf | 29 July 2002 | Medusae | 33469 | Identical to T. NZ1 |
| Tasmania | Turritopsis rubra | Australia, Tasmania, Hobart | 8 June 2004 | Medusae | 34180 | AM183134 |
| Japan Small 1 | Turritopsis dohrnii | Japan Okinawa Island | Early March 2003 | Polyps | – | – |
| Japan Small 2 | Turritopsis dohrnii | Japan | 7 November 2002 | Polyps | – | – |
| Italy 1 | Turritopsis dohrnii | Mediterranean, Italy, | Polyps | – | – | |
| Japan Small 3 | Turritopsis dohrnii | Japan, Okinawa Island | Early March 2003 | Polyps | – | – |
| Italy 2 | Turritopsis dohrnii | Mediterranean, Italy, | 7 November 2002 | Polyps | – | – |
| Italy 3 | Turritopsis dohrnii | Mediterranean, Italy, | 7 November 2002 | Polyps | – | – |
| Italy 4 | Turritopsis dohrnii | Mediterranean, Italy, | 7 November 2002 | Polyps | – | – |
| Italy 5 | Turritopsis dohrnii | Mediterranean, Italy, | 7 November 2002 | Polyps | – | – |
| WHOI 1 | Turritopsis nutricula | USA, Woods Hole; | October 2001 | Polyps | – | – |
| WHOI 2 | Turritopsis nutricula | USA, Woods Hole; | October 2001 | Polyps | – | – |
| Japan Big 1 | Turritopsis pacifica | Japan –Fukushima Prefecture | 2002 | Medusae | – | – |
| Japan Big 2 | Turritopsis pacifica | Japan–Fukushima Prefecture | 2002 | Medusae | – | – |
| Japan Small 4 | Turritopsis small Japanese | Japan - Kagoshima, Kyushu | 6 November 2002 | Medusae | – | – |
| Japan Small 5 | Turritopsis small Japanese | Japan - Kagoshima, Kyushu | 6 November 2002 | Medusae | – | – |
| Japan Small 6 | Turritopsis small Japanese | Japan - Kagoshima, Kyushu | 6 November 2002 | Medusae | – | – |
| Japan Small 7 | Turritopsis small Japanese | Japan - Tanabe Bay | 18 July 2003 | Medusae | – | – |
| Bougainvillia | Bougainvillia spec. | Misaki – Japan | July 2001 | Polyps | – | – |
| Merona | Merona spec. | Mediterranean, off Banyuls-sur-Mer, 62 m | 15 May 2002 | Polyps | – | – |
| Rathkea | Rathkea octopunctata | New Zealand, Hauraki Gulf | 3 July 2002 | Medusae | 33454 | – |
Total genomic DNA was extracted from the ethanol-preserved specimens (polyps or medusae) following an adapted version of the protocol described by Oakley and Cunningham (2000) or Schuchert (2005). The target gene (mitochondrial 16S) was then amplified using the polymerase chain reaction (PCR) and primers SHA and SHB (Cunningham and Buss 1993). The PCR product was purified on a QIAquick spin-column purification kit (Qiagen Inc., Valencia, CA, USA). The purified PCR product was run on a 2% agarose gel stained with ethidium bromide to assess the quantity and quality (i.e. accessory bands) of the product. The purified PCR product (625 base pair long) was then used as a template for double-stranded sequencing using Big Dye as terminator nucleotides. Sequences were read by: the ABI-377 or 3700 automated sequencer (Perkin-Elmer, Wellesley, MA, USA) at Duke University (USA), ABI-377 automated sequencer (Perkin-Elmer) at the Muséum d'Histoire Naturelle (Switzerland) and ABI PRISM 310 automated sequencer (Perkin-Elmer) at the Università di Lecce (Italy). Sequences were first assembled and edited using the software SEQUENCHER 3.0 (Gene Codes Corp., Ann Arbor, MI, USA). They were then aligned using ClustalX (Thompson et al. 1997). All alignments were confirmed and edited by eyes in McClade (Maddison and Maddison 2000). Phylogenetic analyses of the aligned sequences were carried out using PAUP* version 4.0b10 for Macintosh (Swofford 2001). Phylogenetic analyses (heuristic searches) were performed under maximum parsimony and maximum likelihood optimality criteria. Clade stability was assessed by bootstrap analysis (Felsenstein 1985) (100 replicates). The evolutionary model for performing the maximum likelihood analysis was selected using the hierarchical criterion as implemented in Modeltest 3.06 (Posada and Crandall 1998). The model chosen by Modeltest was GTR + I + G.
Results and Discussion
The tree was rooted using three outgroups: Meronasp. (family Oceaniidae like Turritopsis), Bougainvillia sp. (family Bougainvilliidae) and Rathkea octopunctata (Rathkeidae). All the three outgroup families and the ingroup belong to the suborder Filifera. Analyses (parsimony and ML, and parsimony and ML bootstrapping) were run with all three, two (Bouganvillia and Merona) or one (Bougainvillia only or Merona only) outgroup taxa.
The phylogenetic analyses fully support the genus Turritopsis as a monophyletic clade (bootstrap value = 100). Because other genera exist within the family Oceanidae, it is possible that sampling those genera can change our assessment of Turritopsis monophyly.
Topologies of the trees resultant from the various analyses were congruent irrespective of outgroups included in the analysis. Individuals of the genus Turritopsis are clustered in six reciprocally monophyletic clades that correspond to six geographical regions (1, 2). The six clades are: Turritopsis rubra from New Zealand and Tasmania, Turritopsis pacifica (big morph) from northern Japan, Turritopsis (small morph) from southern Japan, Turritopsis dohrnii from Italy and Mallorca, Turritopsis sp. from southern Spain (Mediterranean, Alborean Sea, Andalucia) and T. nutricula from the North-Western Atlantic.
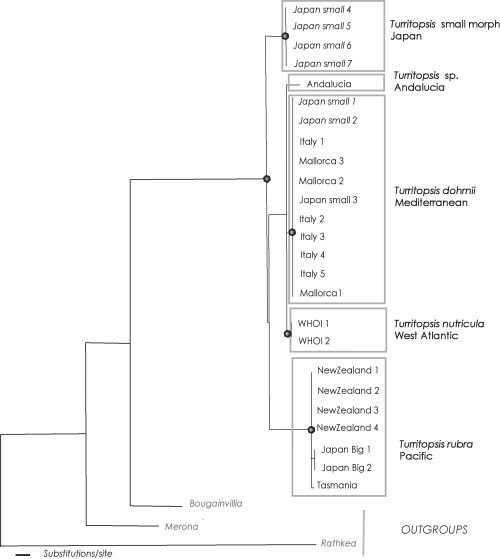
Maximum likelihood tree. In black the nodes within the ingroup that have a bootstrap support >99
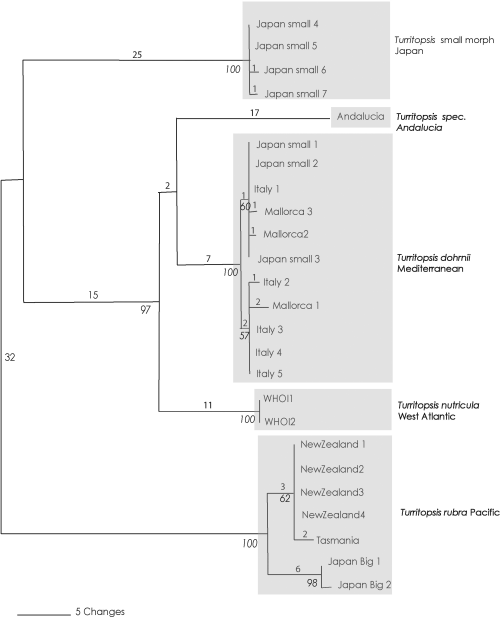
Maximum parsimony tree. Number of changes are indicated above each node. In italic bootstrap values (indicated only if >60)
Very little sequence variation was observed within the individual clades. The samples from New Zealand, originating from localities that are more than 500 km apart, produced all the same sequence. Although more samples must be analysed, there seems to be a quite low intrapopulation variability of 16S (see Govindarajan et al. 2005a; Schuchert 2005 for values of other hydrozoans). Only the Turritopsis dohrnii clade shows some intrapopulation variability with two identifiable clades separated by a difference of only two bases. The T. dohrnii samples came from Italy and Spain (Mallorca), but their sequences did not show geographical patterning. Instead, the identified clades contain sequences from both sampling regions. The divergence of the two clades is quite low and it seems unlikely they represent two separate species.
The Mediterranean Turritopsis are clearly separate from T. nutricula of the Eastern Atlantic, thus confirming the opinion of Schuchert (2004). The Mediterranean clade could clearly be ascribed to T. dohrnii because enough life cycle information is available for several samples (the Balearan samples are described in Schuchert 2004). Samples of these two species form two highly supported clades (bootstrap = 100). The sister group to T. dohrnii appears to be the distinct Turritopsis from southern Spain (Alborean Sea) revealing a Mediterranean clade (bootstrap support = 100). The Turritopsis from southern Spain was represented only by a small polyp colony and, therefore, it was impossible to identify it reliably at the specific level. Because it was found not far from the Gibraltar Strait, it may represent Turritopsis polychirra (the only Turritopsis species of the North-Eastern Atlantic). Unfortunately, we were unable to procure DNA samples of definitive Turritopsis polycirrha and test this assertion. However, the small size of the colony and the proximity of the sampling site to the Atlantic argue much in favour of this hypothesis. Regardless, the 16S sequence of the Turritopsis specimen from southern Spain is quite distinct from those obtained from T. dohrnii specimens and the West Atlantic T. nutricula specimens (Fig. 2).
Based on morphological and life history differences, Schuchert (2004) regarded the Turritopsis from New Zealand as distinct from T. nutricula of the Eastern Atlantic. The name T. rubra (Farquhar 1895) was therefore revived for the New Zealandic population. The present molecular results (Fig. 1) clearly support this view and T. rubra must be seen as a valid species. Our analysis included a polyp colony from the type locality of T. rubra (Wellington Harbour), and it yielded the same haplotype found in medusae originating from Auckland. This indicates that T. rubra is the only representative of this genus in New Zealand. A Turritopsis Medusa from Tasmania, not differing morphologically from T. rubra, also falls in the T. rubra clade, expanding the known range of the species.
Despite the fact that all Japanese Turritopsis are usually assigned to T. nutricula, sequences of the Japanese Turritopsis split into three well-separated groups (1, 2. Two morphs of Turritopsis were identified in Japan based on a recent morphological analysis: a northern population with large medusae and tentacles arranged in two rows, and a southern population with much smaller medusae and only one row of tentacles (Kubota 2005). One clade of Japanese Turritopsis corresponds to the big morph, (from northern Japan) is likely attributable to T. pacificaMaas 1909 (see taxonomic part in Table 2 for more details on this nominal species). Our results clearly show that T. pacifica is a distinct species that is separate from T. nutricula of the Western Atlantic. Turritopsis pacifica forms a well-supported clade, but its divergence from T. rubra from New Zealand and Tasmania is small and only in the maximum likelihood analyses they sorted not reciprocally monophyletic (1, 2). We are thus unable to make definite decisions on the species status of T. pacifica. A more detailed analysis including a larger sequence data set from the eastern Pacific is needed to evaluate whether T. pacifica is conspecific with T. rubra or an independent species. More information on its reproductive strategies is also needed to permit a conclusive answer (see also Table 2).
| Species | Recent references | Type locality | Distribution | Colony characters | Adult medusa characters | Remarks |
|---|---|---|---|---|---|---|
| Turritopsis nutricula McCrady, 1857 | Kramp (1961), Wedler and Larson (1986), Calder (1988),Migotto (1996), Schuchert (2004) | Charleston Harbour, SC, USA | From New England to Brazil, according to Mayer (1910) common in the Bahamas and the Tortugas | Erect, branching hydroid colonies. Periderm two-layered. Hydranths spindle- to club-shaped; filiform tentacles scattered over hydranth body. Gonophores on the hydrocauli in perisarc-covered region | Up to 6 mm in height, 40–100 tentacles, tentacle tips can be swollen, vacuolated cells in four distinct masses. Sexes separate, females oviparous. Colour: manubrium dull-yellow or orange, ocelli dark-brown or orange. Egg diameter 0.116 mm | The restriction of the distribution to the western Atlantic takes account of the revision of Schuchert (2004) and the molecular results obtained in this study |
| Turritopsis fascicularis Fraser, 1943 | Fraser (1943), Fraser (1944) | 24.70°N 80.46°W, 215 m | Known from the type locality only | Polysyphonic | Unknown | Fraser (1944) distinguished this species from T. nutricula on account of its polysiphonic colonies. Calder (1988) regarded it as distinct from T. nutricula and perhaps conspecific with T. dohrnii |
| Turritopsis polycirrha (Keferstein, 1862) | Kramp (1928), Russell (1953), Schuchert (2004) | St Vaast, Normandy, France | English Channel; Great Britain, up to Firth of Forth in the east, up to Bristol Channel in the west; southern parts of the North Sea, sometimes as far east as Helgoland | Polyp phase inadequately known from nature, likely a stolonal or only sparingly branched colony, hydranths with scattered filiform tentacles, hydranth colour: red | Adult medusa 4–5 mm in height and diameter, 80–90 tentacles with tips not inflated, manubrium without gelatinous peduncle and on top four blocks of vacuolated cells fused into a single compact mass, radial canals overtop this mass. Vacuolated cells continued downward on manubrium as bulging, perradial ribs. Radial canals broad. Simultaneous hermaphrodites and larviporous. Colour: stomach and gonads brilliant red to dark crimson | This species is kept separate from T. nutricula primarily based on its larviparity and hermaphroditism, but there are also morphological differences |
| Turritopsis dohrnii Weismann, 1883 | Ramil and Vervoort (1992), Piraino et al. (1996), Carla et al. (2003), Schuchert (2004) | Naples, Mediterranean | Western Mediterranean, Adriatic Sea, Japan | Hydroid colony of variable height, either sparingly branched with shoots only a few mm high to much branched and polysiphonic colonies up to 35 mm high. Hydranths with 12–20 tentacles. Hydranths in life colourless or pinkish | Up to 2.7 mm in height, diameter 3.2 mm, 14–32 tentacles, manubrium reaching to bell margin, tentacles sometimes with terminal swellings, ocelli rust-coloured, gonads brownish, with four interradial rust-coloured dots, proximal ends of radial canals swollen through vacuolated gastrodermal cells, the four swellings not fused into a single mass. Sexes separate and females oviparous. Medusa can metamorphose back into polyp stage | This species was originally based on the hydroid stage. Schuchert (2004) regarded it as a distinct species, though very similar to T. nutricula. The molecular results of this study strongly support this view |
| Turritopsis chevalense (Thornely, 1904) | Schuchert (2003) | Cheval Paar, Gulf of Manaar, Ceylon, 11–15 m on shells of Pecten and sea weeds | Tropical Indian Ocean, ? Indonesia | Colonies branched, with adnate pedicels bearing medusae buds | Unknown | The colonies of Corydendrium chevalense are almost certainly referable to the genus Turritopsis. No information on the adult medusa is available, and T. chevalense is at present inadequately known and not reliably identifiable |
| Turritopsis pleurostoma (Péron and Lesueur, 1810) | Haeckel (1879) | De Witt's Land, Western Australia | Only known from type locality | Unknown | Medusa is about 30–40 mm high, has 32 tentacles and reddish-brown gonads | The very large size and the low number of tentacles do not match any other Turritopsis medusa and therefore the species is here regarded as possibly valid. Some doubts remain because 3–4 cm appear very large for a Turritopsis medusa. Apart from its first description, it has never been reported anymore |
| Turritopsis rubra (Farquhar, 1895) | Ralph (1953), Schuchert (1996, 2004) | Wellington Harbour, New Zealand | New Zealand, Tasmania | Colonies from almost stolonal to much branched, polysiphonic reaching 5 cm in height. Hydranths with up to 20 tentacles, periderm composed of two layers. Young medusae released with eight tentacles, with orange interradial pads on manubrium | Adult medusa up to 7 mm in height, usually 3–4 mm, umbrella top conical or flat, up to 120 tentacles in one or two rows depending on state of contraction, tentacle tips only very slightly swollen, on top of manubrium four masses of vacuolated cells fused into a disc-like cushion, radial canals reaching nearly top of subumbrella. Sexes separate, females larviparous, planulae may even transform into primary polyps on manubrium. Colour: stomach and gonads brilliant red | The Tasmanian specimens observed were morphologically indistinguishable from the New Zealandic T. rubra. The 16S DNA sequences in the present work showed three base substitution (0.5%). As the Mediterranean T. dohrnii has a maximal intraspecific variation of about 1% (Fig. 1), the difference between the New Zealandic and Tasmanian samples are thought to represent intraspecific variation |
| Turritopsis lata von Lendenfeld, 1885 | Blackburn (1937) (hydroid), Watson (1978) | Port Jackson (Sydney Harbour), New South Wales, Australia | Known from the type locality only | Up to 3.5 mm high, short peduncle not covered by vacuolated cells, vacuolated cells not forming a continuous mass, confined to proximal part of radial canals and along peduncle. Stomach spherical to spindle shaped, gonads in four ovoid pads. Tentacle number in subadults or adults: 20–130. Gonads intensively brown | The available data suggest that T. lata is distinct from other Turritopsis species. Examination of new material from the type locality is needed for a further evaluation of T. lata | |
| Turritopsis pacifica Maas, 1909 | Yamada and Nagao. (1971),Hirohito Emperor of Japan (1988),Park (1990), Kubota et al. (2003), zSchuchert (1996, 2004) | Misakai, Sagami Bay, Japan | Japan | Adult medusa 8–9 mm in height, 120–150 tentacles usually in two rows (may depend on state of contraction), four masses of vacuolated cells fused to one block. Sexes presumably separate. Colours: gonads orange to red | The existing descriptions of T. pacifica portray this species as indistinguishable from T. rubra (comp. Schuchert 1996, 2004). It is unclear whether T. pacifica is gonochoristic or larviparous. Preliminary observations (S. Kubota, unpublished data) indicate that it is larviparous like T. rubra. The present molecular data suggest that they could be conspecific | |
| Turritopsis minor (Nutting, 1905) | Cooke (1977) | Albatross stations 3859, 4077 and 4098, Islands of Molokai and Maui, Hawaiian Archipelago | Known from the type locality only | Because no life-cycle information is available, it must remain an insufficiently characterized species. The Hawaiian hydroid identified as T. nutricula by Cooke (1977) is here referred to as T. minor for biogeographical reasons |
The second clade of Japanese Turritopsis is found exclusively in southern Japan (identified as ‘small morph’, because it forms a much smaller Medusa than T. pacifica) and forms a very well-supported clade. This species is very similar to the Mediterranean T. dohrnii in life cycle characteristics, as well as in polyp and Medusa morphology. However, it is genetically distinct from T. dohrnii and it probably represents an as yet unnamed species (for a detailed report on the geographical distribution of the two Japanese morphs of Turritopsis see Kubota 2005). The aforementioned clade does not comprise all medusae of the small type found in Japan. Surprisingly, the sequences of two medusae coming from Japan and identified as Turritopsis‘small morph’ as well as a polyp colony (see Table 1) proved to be identical to one of the Italian T. dohrnii sequences. In order to rule out any confusion, contamination or mislabelling of material, this result was confirmed by sequencing independent samples of the Japanese Turritopsis‘small morph’ in two different laboratories (University of Lecce, Italy and Duke University, USA). The presence of a Mediterranean haplotype of T. dohrnii in Japan is best explained by a relatively recent introduction in southern Japanese waters (because the Mediterranean samples produced several different haplotypes and the Japanese not, we assume this direction of transport and not the reverse). This recent introduction is most likely due to human activities, e.g. by ballast water of cargo ships. Turritopsis dohrnii is able to revert their life cycle, usually through formation of temporary resting stages, that can much better resist unfavourable conditions than the Medusa stage (Bavestrello et al. 1992; Piraino et al. 1996, 2004). Therefore, the possibility of being transported by human activities appears even more likely.
The topology of the tree obtained for the 16S sequences (1, 2) invites some speculations on the origins of the genus Turritopsis. All Atlantic-Mediterranean sequences form a monophyletic clade nested within the Pacific sequences (with exception of the T. dohrnii samples that were probably introduced into Japan recently). This pattern could be explained by regarding the Atlantic Turritopsis as being derived from a trans-arctic migration event. Many intertidal species are thought to have entered the North Atlantic from the North Pacific about 3.5 mya when the Arctic Ocean was warm enough to allow boreal and temperate species to migrate between the North Atlantic and Pacific (Vermeij 1991).
Although the analysis of species boundaries in the genus Turritopsis as presented here is still far from being complete, our results show that 16S sequences are very useful for tackling difficult systematic questions at the species level within Hydrozoa, a conclusion also reached by Govindarajan et al. (2005a,b) and Schuchert (2005).
More sequences are needed from all populations, especially those representing Turritopsis polycirrha and T. chevalense that could not be obtained for this study. A more complete sampling will allow for the generation of the phylogenetic framework necessary for a comprehensive revision of the genus.
From an evolutionary point of view the genus Turritopsis shows some unusual life-history features that are uncommon within the Hydrozoa. Some species or populations brood their larvae (Schuchert 2004) and others are able to revert their life cycle (Piraino et al. 1996). Because not all forms show these traits, it would be interesting to track their evolutionary origin using a phylogenetic hypothesis. These interesting traits may have originated in a single event or multiple times due to a common selective pressure.
Further studies taking into consideration these features may contribute to a better understanding of the evolution of life cycle strategies in the Turritopsis and in the whole Hydrozoa taxon.
Acknowledgements
We wish to thank the Cunningham laboratory at Duke University (NC, USA) where most of the molecular work was done. This work was greatly improved by comments and suggestions by Allen Collins.
We profited very much from a gift of Turritopsis material from Tasmania provided by Dr L. Turner of the Tasmanian Museum and would like to express our gratitude for this.
This work was supported by the National Science Foundation PEET Grant No. DEB-9978131 A000, and by the MURST (COFIN and FIRB Projects), the Administration of the Province of Lecce, the ICRAM (Project ‘Identificazione e distribuzione delle specie non indigene nel Mediterraneo’) and the European Union (Marie Curie Contract No. HPMD-CT-2001-00099, and the MARBEF network).




