Variation at the insulin-like growth factor 1 gene and its association with body weight traits in the chicken
Summary
The insulin-like growth factor 1 (IGF1) is essential for normal embryonic and postnatal growth in mammals. In this study, a total of 342 F2 individuals, derived from Broiler crossing to Baier layer (Northeast Agricultural University Resource Population, NEAURP), were used to investigate the associations of haplotypes in the chicken IGF1 (cIGF1) gene with body weight traits. Primers for the 5′-flanking, exon 3 and 3′-flanking regions of cIGF1 were designed according to chicken genome database. Single nucleotide polymorphisms (SNPs) between parental lines were detected by sequencing, and PCR restriction fragment length polymorphism (PCR-RFLP) and PCR single-stranded-conformation polymorphism (PCR-SSCP) methods were used to genotype the SNPs in the population. Haplotypes were constructed with the three SNPs detected. The association analysis showed that haplotypes based on three cIGF1 polymorphisms (c.-366A>C, c.528G>A and c.*1024C>T) were associated with body weight traits, suggesting that cIGF1 or a tightly linked gene had effects on body weight in the chicken.
Introduction
The insulin-like growth factor system is a complex system of peptide hormones (IGF1 and IGF2), cell surface receptors and circulating binding proteins. IGF1 and IGF2 bind to the insulin-like growth factor 1 receptor (IGF1R), insulin receptor (IR) and activate their intrinsic tyrosine kinase domain activities (Adam et al. 2005). These activated receptors initiate signal cascades and ultimately result in regulation of a number of biological responses (Adam et al. 2005). These components act together to control a number of crucial biological outcomes including cellular growth, proliferation, differentiation, survival against apoptosis and migration (Khandwala et al. 2000; Pollak et al. 2004). These processes are related to tissue formation and remodelling, bone growth, brain development and energy metabolism, which all ultimately influence organism size and longevity.
The chicken IGF1 gene has been cloned and sequenced (Kajimoto & Rotwein 1991) and is composed of four exons and three introns, spanning over more than 50 kb on chromosome 1. The mature cIGF1 with a molecular weight (MW) of 7 kDa is a non-glycoprotein hormone whose structure is homologous to pro-insulin (Humbel 1995). Plasma cIGF1 concentration is greater in genetic lines selected for high growth rate compared with that in slower growing lines (Scanes et al. 1989). Plasma cIGF1 concentration and hepatic gene expression increase rapidly with aging after hatching, reach a peak before sexual maturity, and then decline (Kikuchi et al. 1991). Moreover, there is ample evidence suggesting that cIGF1 might influence growth rate, body composition and lipid metabolism in poultry (McMurtry 1998; Tomas et al. 1998; Beccavin et al. 2001). Seo et al. (2001) reported the relationship of the polymorphic sites to body weight and cIGF1 concentration of male Korean Ogol chicken. It was found that the Pst I RFLP was associated with BW at 2 and 4 months of age in the Wanzhai Yellow breeds (Wang et al. 2004). The c.*1024C>T in the 3′-flanking region of cIGF1 was associated with growth and carcass traits including body weights at hatch and 12 weeks of age (BW0 and BW12), carcass weight (CW) and abdominal fat weight (AFW; Lei et al. 2005). PCR-RFLP was developed to genotype a chicken F2 population and to evaluate associations between each SNP genotype and multiple phenotypes. Significant associations (p < 0.0125) were found between cIGF1 and 5-week body weight (Bennett et al. 2006). The c.-828C>T SNP of cIGF1 was significantly linked with the transversal area of the leg muscle fibre and transversal area of the breast muscle fibre (Lei et al. 2007).
To help interpret the genetic control of growth in chickens, cIGF1 was examined as a candidate gene for body weight traits. The objectives of the current study were to identify SNPs of cIGF1, develop PCR-RFLP and PCR-SSCP methods to detect those DNA polymorphisms in a unique chicken F2, and evaluate associations between the variation at cIGF1 and that of body weight traits.
Materials and methods
Experimental population and management
A two-generation resource family, Northeast Agricultural University Resource Population (NEAURP), was used in the current study. The NEAURP was established by crossing broiler sires, derived from Northeast Agricultural University broiler lines divergently selected for abdominal fat content, with Baier layer dams, a Chinese local breed. The lean and fat broiler lines originally derived from a commercial Arbor Acres grandsire line, and grandsires of the NEAURP were from fat line. The Baier layer is a Chinese local egg-laying breed that has yellow feathers, a yellow beak, yellow feet and white ears. Granddams of the NEAURP were from the Baier breed. The F1 birds (12 sires and 72 dams) were intercrossed to produce the F2 population (Wang et al. 2006). A total of 342 F2 individuals from four F1 sires were used in the current study. All birds had ad libitum access to feed and water. Commercial corn–soybean-based diets that met all National Research Council (1994) requirements were provided in the study. From hatch to 3 weeks of age, birds received a starter feed (3100 kcal of ME/kg and 210 g/kg of CP) and from 3 to 12 weeks of age, birds were fed a grower diet (3000 kcal of ME/kg and 190 g/kg of CP; Wang et al. 2006).
Animal care and handling were conducted in accordance with policies on the care and use of animals promulgated by the ethical committee at NEAU.
Phenotypic measurements
Body weights (BW) were measured at hatch and weekly up to 12 weeks of age, carcass weight (CW) was measured at 12 weeks of age. Statistics of body weight traits is shown in Table 1.
| Traits | NEAURP | Baier layers | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
 1
1 |
SD2 | CV3 | Min4 | Max5 | 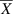 1
1 |
SD2 | CV3 | Min4 | Max5 | |
| BW0 (g) | 39.43 | 3.20 | 7.65 | 32.30 | 47.05 | 32.14 | 2.90 | 9.04 | 25.76 | 97.15 |
| BW2 (g) | 169.10 | 20.82 | 12.31 | 119.90 | 235.00 | 93.39 | 20.53 | 21.99 | 40.00 | 140.00 |
| BW4 (g) | 461.66 | 73.29 | 15.87 | 265.00 | 660.00 | 226.14 | 48.51 | 21.45 | 95.00 | 310.00 |
| BW6 (g) | 844.52 | 124.84 | 14.78 | 515.00 | 1155.00 | 420.60 | 94.06 | 22.36 | 195.00 | 550.00 |
| BW8 (g) | 1284.18 | 208.24 | 16.22 | 810.00 | 1845.00 | 622.23 | 150.14 | 24.13 | 330.00 | 860.00 |
| BW10 (g) | 1709.73 | 286.03 | 16.73 | 1100.00 | 2520.00 | 802.23 | 192.46 | 23.99 | 475.00 | 1130.00 |
| BW12 (g) | 2078.08 | 368.43 | 17.73 | 1075.00 | 3020.00 | 985.47 | 216.55 | 21.97 | 610.00 | 1375.00 |
| CW (g) | 1838.06 | 332.94 | 18.11 | 960.00 | 2730.00 | 868.14 | 208.18 | 23.98 | 540.00 | 1215.00 |
-
1
 , mean; 2SD, standard deviation; 3CV, coefficient of variation (%); 4Min, minimum value; 5Max, maximum value. NEAURP, Northeast Agricultural University Resource Population. BW, body weight; CW, carcass weight.
, mean; 2SD, standard deviation; 3CV, coefficient of variation (%); 4Min, minimum value; 5Max, maximum value. NEAURP, Northeast Agricultural University Resource Population. BW, body weight; CW, carcass weight.
DNA, RNA extraction and cDNA synthesis
Genomic DNA was extracted by phenol–chloroform method from 10 μl venous blood collected in EDTA-Na2-coated tubes. The RNA was extracted from liver with the Trizol reagent (Invitrogen, Rockville, MD, USA) according to the instructions of the manufacturer, then dissolved in diethyl pyrocarbonate-treated water. The reverse transcription reactions were carried out using the Takara RNA PCR Kit (Version 3.1, Takara Biotechnology Co. Ltd., Dalian, China). The reverse transcription reaction contained 1 μg of total RNA derived from the liver tissue of two broilers and two Baier layers in a final volume of 20 μl, respectively.
Sequence variation of cIGF1
Primers for the coding region of cIGF1 (5′-CTC TAA TCC CTC TTC TG-3′; 5′-ACA TTC TAG TAC GGT AG-3′) that amplify a 669-bp fragment were designed using Primer premier 5.0 (Premier Biosoft International, Palo Alto, CA, USA) according to the sequence in the GenBank database (accession number M32791). Four individuals, two broiler and two Baier birds, were selected to detect SNPs by sequencing the coding region. The PCR reaction system included 1 × PCR reaction buffer (10 mm Tris–HCl, 50 mm KCl and 1.5 mm MgCl2, pH 8.3), 0.3 mm dNTP, 5 pmol of each of the forward and reverse primers, 1 μl of cDNA, and 1 U of Taq DNA polymerase (Takara Biotechnology Co. Ltd) in a final volume of 25 μl. The PCR conditions were 94°C for 5 min, followed by 35 cycles of 94°C for 30 s, 55°C for 30 s, 72°C for 1 min, with a final extension step of 72°C for 10 min. The PCR products were cloned into pMD 18-T vector (Takara Biotechnology Co. Ltd). Then, the ligated products were transformed into Escherichia coli DH5α and sequenced in forward and reverse directions with an ABI 3730 sequencer (Bioasia Biotechnology Co., Ltd. Shanghai, China). Restriction enzyme sites in these sequences were predicted by Primer Premier 5.0.
Genotyping of the cIGF1 gene in the F2 population
Three pairs of primers for 5′-flanking region (5′-CAC AGC CAC CCG AAA GT-3′; 5′-AGA AAT CAC AAA AGC AGC AC-3′), exon 3 region (5′-TAC ACA TCT ACC ACT GTC AT-3′; 5′-TCC TCA GGT CAC AAC TCT-3′) and 3′-flanking region (5′-GTA CAA CGG TGC TAT TT-3′; 5′-AGG ACA CTG TTG GCT AT-3′) that, respectively, amplify 542, 209, 746-bp fragment, were designed using Primer premier 5.0 (Premier Biosoft International) according to chicken genomic DNA sequence in the UCSC Genome Bioinformatics site (http://genome.ucsc.edu.cn). The 10-μl reaction volume included 50 ng of template, 1 × reaction buffer, 2 pmol of each of the forward and reverse primers, 0.16 mm dNTP, 1.5 mm MgCl2 and 0.5 U of Taq polymerase. The PCR conditions were the same as described above.
A PCR amplification of DNA from each F2 bird was performed as previously described. The 542 or 746-bp PCR product were separately digested with 3 U of restriction enzyme Hinf I (Takara Biotechnology Co. Ltd) and Bsp119 I (Sangon Biological Engineering Technology & Services Co. Ltd. Shanghai, China) at 37°C overnight. The restriction digests were run in electrophoresis for 0.5 h at 120 V on a 2.0% agarose gel with ethidium bromide. Individual PCR-RFLP fragment sizes for the gene were determined by visualizing the band pattern under ultraviolet light.
The 209-bp PCR product was detected by PCR-SSCP method. One microlitre of the PCR products was mixed with 5 μl of loading buffer (98% formamide, 0.025% bromophenol blue, 0.025% xylene cyanol, 10 mm EDTA and 10% glycerol), then the mixture was denatured in 98°C for 10 min, and placed on ice for 5 min. Finally, the mixture was run in electrophoresis for 18 h at 10 V/cm on a 14% polyacrylamide gel. Silver stain method was developed to display the bands. Individual PCR-SSCP band patterns were determined under visible light.
Haplotype construction and linkage disequilibrium analysis
In this study, haplotypes were constructed based on three SNPs in 342 experimental animals by Phase 2.1 program (Stephens et al. 2004).
Linkage disequilibrium (LD) is the non-random association of alleles at adjacent loci (Kristin et al. 2002). Two normalized measures, D′ and r2, were used to characterize LD patterns within the studied candidate gene. Pair-wise D′ and r2 values were computed (Lewontin 1964; Devlin & Risch 1995). The values of |D′| > 0.33 and r2 > 0.1 were applied as the criteria for meaningful LD (Kruglyak 1999; Moffatt et al. 2000; Nakajima et al. 2002).
Statistical Analysis
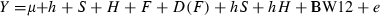
Results
Sequence variation and polymorphisms analysis
The c.-366A>C was found at the 366 bp 5′ of the ATG translation initiation codon of cIGF1 (Amills et al. 2003). The Hinf I-digested PCR products had a fragment of 542 bp for the CC genotype, 428 bp for the AA genotype and a combination of 542 and 428 bp for the AC genotype. The aligned complete coding sequences revealed a G/A substitution in exon 3 of cIGF1. This mutation was defined as c.528G>A in the present study. The PCR-SSCP method was developed successfully for genotyping the SNP. Three genotypes were detected and designated as AA, AG and GG. The c.*1024C>T was found at the 1054 bp 3′ of the translation stop codon of cIGF1 (Lei et al. 2005). The Bsp119 I-digested PCR products had a fragment of 746 bp for the TT genotype, 527 bp for the CC genotype and a combination of 746 and 527 bp for the CT genotype.
Linkage disequilibrium and haplotype – trait association analysis
In the current study, there were seven haplotypes in the population compared with possible eight haplotypes. Five main haplotypes with the minor frequency of above 1% accounting for 98.8% of the observations were used in haplotype – trait association analysis (Table 2). The linkage disequilibrium analysis indicated that c.-366A>C and c.528G>A (r2 = 0.49, |D′| = 0.98), c.528G>A and c.*1024C>T (r2 = 0.13, |D′| = 0.41), as well as c.-366A>C and c.*1024C>T (r2 = 0.43, |D′| = 0.99) were all in LD. The haplotypes of cIGF1 were suggestively associated with BW at 4 and 12 weeks age and CW (p < 0.05). The haplotypes of cIGF1 were significantly associated with BW at 2, 6, 8 and 10 weeks of age (p < 0.0071). There were significant differences between AGT and CAC haplotype in BW at 2–12 weeks of age and CW (p < 0.05) (Table 2).
| Trait1 | P-value2 | AGT (3283) | CGC (68) | CAC (144) | CGT (46) | CAT(90) |
|---|---|---|---|---|---|---|
| BW2 (g) | 0.0051 | 171.3 (3.24)a | 165.3 (3.9)b | 165.6 (3.4)b | 171.1 (4.1)ab | 170.2 (3.6)ab |
| BW4 (g) | 0.0245 | 467.7 (5.7)a | 453.0 (8.7)ab | 450.4 (6.7)b | 461.4 (10.2)ab | 462.7 (7.9)ab |
| BW6 (g) | 0.0003 | 859.2 (10.0)a | 832.7 (15.5)ab | 815.0 (11.8)b | 836.4 (18.2)ab | 855.2 (14.0)a |
| BW8 (g) | 0.0006 | 1311.2 (17.9)a | 1265.9 (25.9)bc | 1245.1 (20.3)c | 1280.4 (30.6)abc | 1305.1 (23.7)ab |
| BW10 (g) | 0.0002 | 1745.2 (21.5)a | 1678.3 (32.7)bc | 1652.8 (25.0)c | 1700.7 (39.2)abc | 1731.5 (29.8)ab |
| BW12 (g) | 0.0147 | 2109.9 (27.7)a | 2041.7 (41.2)ab | 2028.6 (32.0)b | 2083.2 (47.9)ab | 2108.5 (37.2)a |
| CW (g) | 0.0149 | 1864.8 (26.2)a | 1800.0 (37.8)bc | 1793.6 (29.8)c | 1846.5 (435.7)abc | 1864.5 (34.4)ab |
- 1BW, body weight (weeks 2–12); CW, carcass weight (12 wk).
- 2Comparison of haplotypes within a trait.
- 3Individual number in parentheses.
- 4Standard error in parentheses.
- a–cMeans within a row with no common superscript differ significantly by contrast tests, p < 0.05.
Discussion
As a traditional approach for studying both trait association (marker versus trait) and LD (marker versus marker), single marker analysis has created many problems, such as time consuming and lower efficiency to detect QTL (Daly et al. 2001). A disadvantage of performing an association study in a temporally variable population is that spurious associations may be generated for any trait and locus that are under selection regardless of whether the locus has a causal effect on variation in the trait (Morsci et al. 2006). The analysis based on haplotype or haplotype block provides a practical solution to resolve these problems (Ott & Rabinowitz 1997; Daly et al. 2001) and has greater statistical power by linkage information (Jenny et al. 2004; Li et al. 2004). In the current study, haplotypes were constructed with the three SNPs and analysed for finding the association with body weight traits. The pairs-wise r2 and D′ analysis showed that the three SNPs were in LD. The association analysis showed that the gene haplotypes were suggestively associated with BW at 4, 12 weeks of age and CW (p < 0.05). Haplotypes had significant effects on BW at 2, 6, 8 and 10 weeks of age (p < 0.0071). The effect (least-square mean) of AGT haplotype was significantly larger than that of CAC haplotype. The AGT haplotype was associated with increased BW and CAC with reduced BW. It was concluded that AGT was the advantageous haplotype and CAC was unfavourable for BW.
Among the three SNPs, the c.-366A>C had been reported that it was involved in the suppression of one potential CdxA transcription factor-binding site (Amills et al. 2003). The birds with CdxA-binding allele A increased the transcription efficiency of cIGF1 in the small intestine (Yan, China Agricultural University, personal communication). Additionally, it had been reported that there were significant associations between c.-366A>C and growth and body weight traits in other independent chicken resource population (Amills et al. 2003; Zhou et al. 2005; Li, China Agricultural University, personal communication). The c.528G>A mutant does not involve any amino acid change, and may be linked with some functional sites in cIGF1. The c.*1024C>T had been reported that it was associated with BW0, BW12 and CW by least square analysis in other independent chicken resource population (Lei et al. 2005). Therefore, c.*1024C>T may be a causal site or be closely linked to other gene(s) affecting BW traits in chicken.
The associations revealed through haplotype analysis method might be attributed to the close linkage of three SNPs with BW QTL on chromosome 1. Incidentally, in a broiler layer, F2 population used to map BW QTL by a genome scan, a QTL affecting BW at 6 weeks of age has been found at 160 cM (confidence interval 114–180 cM) covering cIGF1 on chromosome 1 (Sewalem et al. 2002). These findings were strongly indicative of cIGF1 as a candidate gene of QTL for growth in chickens.
Acknowledgements
The authors gratefully acknowledge the members of the Poultry Breeding group of the college of Animal Science & Technology in the Northeast Agricultural University for help in managing the birds and collecting the data. This research was supported by National Natural Science Foundation Key Project (No. 30430510), National Basic Research Programme (No. 2006CB102105) and Programme for New Century Excellent Talents in University (No. NCET - 04 - 0343).




