Relative predispositional effects of HLA class II DRB1-DQB1 haplotypes and genotypes on type 1 diabetes: a meta-analysis
Abstract
The direct involvement of the human leukocyte antigen class II DR-DQ genes in type 1 diabetes (T1D) is well established, and these genes display a complex hierarchy of risk effects at the genotype and haplotype levels. We investigated, using data from 38 studies, whether the DR-DQ haplotypes and genotypes show the same relative predispositional effects across populations and ethnic groups. Significant differences in risk within a population were considered, as well as comparisons across populations using the patient/control (P/C) ratio. Within a population, the ratio of the P/C ratios for two different genotypes or haplotypes is a function only of the absolute penetrance values, allowing ranking of risk effects. Categories of consistent predisposing, intermediate (‘neutral’), and protective haplotypes were identified and found to correlate with disease prevalence and the marked ethnic differences in DRB1-DQB1 frequencies. Specific effects were identified, for example for predisposing haplotypes, there was a statistically significant and consistent hierarchy for DR4 DQB1*0302s: DRB1*0405 =*0401 =*0402 > *0404 > *0403, with DRB1*0301 DQB1*0200 (DR3) being significantly less predisposing than DRB1*0402 and more than DRB1*0404. The predisposing DRB1*0401 DQB1*0302 haplotype was relatively increased compared with the protective haplotype DRB1*0401 DQB1*0301 in heterozygotes with DR3 compared with heterozygotes with DRB1*0101 DQB1*0501 (DR1). Our results show that meta-analyses and use of the P/C ratio and rankings thereof can be valuable in determining T1D risk factors at the haplotype and amino acid residue levels.
Introduction
In the 1970s, association and affected sib pair linkage data established the role of human leukocyte antigen (HLA) in type 1 diabetes (T1D) (1, 2). Although multiple non-HLA genes have been implicated, the HLA region genes are the major T1D susceptibility loci known. By the 1980s, heterogeneity in the HLA component to T1D was well established with a complex hierarchy of predisposing, intermediate, and protective effects at the genotype and haplotype levels (3). A direct role of the HLA class II DRB1, DQA1, and DQB1 genes as the major HLA component to T1D has been established (4–6). Numerous studies have investigated the relative predispositional effects (RPEs) of the HLA DR-DQ alleles, haplotypes, and genotypes (3, 7–17). Cross ethnic studies have been particularly informative in determining both the direct role of HLA DR-DQ genes in T1D and their RPEs (5, 6, 18–23). Also, various HLA DRB1, DQA1 and DQB1 amino acid sites, either individually or in various combinations, have been suggested as T1D components (6, 12, 15–16, 24–28).
Controlling for the influence of class II DR-DQ effects, the role of additional HLA class II (DPB1) (29, 30) and class I genes including age of onset effects and rapid disease progression (31–37) in T1D risk has been shown. Also, the presence of additional disease-predisposing loci on specific high-risk DR-DQ haplotypes, i.e. DRB1*0301-DQB1*0201 (DR3), DRB1*0401-DQB1*0302, DRB1*0404-DQB1*0302 (37–43), and the protective DRB1*1501-DQB1*0602 haplotype (44), has been shown. The results from study of the effects of additional disease genes using different populations and even different samples from similar populations are very heterogenous, reminiscent of non-HLA gene effects (37, 43). The HLA DR-DQ contribution to T1D is by far the strongest genetic effect both within the major histocompatibility complex and across the genome.
HLA allele and haplotype frequencies vary considerably across ethnic groups (45–47) as does T1D risk (48). Asian populations show much lower frequencies of the T1D high-risk DR-DQ haplotypes mentioned above, which are relatively common in Caucasian populations; the disease prevalence in these two ethnic groups mirror these differences. Some HLA genes and amino acids have been shown to be under strong balancing selection at the population level (34, 45, 46, 49, 50). Ultimately, one would like to coordinate disease and population level results more directly at the individual allele, haplotype, genotype, and amino acid levels.
Given the marked ethnic differences in HLA DRB1-DQB1 frequencies, the question we have addressed is the following: Do the DR-DQ haplotypes and genotypes show the same RPEs across all populations and ethnic groups? They should, given the direct role of DR-DQ in disease. If there is consistency across the populations in the RPEs, then they can be used to identify the amino acids involved in T1D. However, if there are disease-predisposing genes in the HLA region additional to DR-DQ, which may well show haplotype-specific effects, plus regional and ethnic variation, then the RPEs may differ. These would be candidates for study of additional disease modifying genes in the HLA region and identification of additional HLA region genetic factors that are involved in T1D.
Our analyses include both comparisons of risk within a population and meta-analyses across populations and ethnic groups. We introduce a new statistic, the patient/control (P/C) ratio of haplotype or genotype frequencies within a study (also allele and amino acid frequencies) that allows comparison of absolute penetrance values within and across studies. Our first step was to determine, within each data set, the RPEs of the HLA DR-DQ haplotypes and genotypes and for some specific haplotypes to identify those that differ significantly in their T1D effects. Although there are genotype-specific effects in T1D, and ultimately these are the effects we want to understand, it is still appropriate to consider haplotype effects (averaged over genotypes). We show below that consistent patterns are seen for most HLA DRB1-DQB1 haplotypes across populations. The total number of haplotypes observed is less than the number of genotypes, so statistical power is higher at the haplotype level. We were only able to study genotype effects in a much reduced number of populations (restricted by availability of the genotype data and sample size), and statistically significant results could only be tested for in a limited set of genotypes.
Study populations and methods
Study populations
Notation: Because of the varying definition of HLA typing in various studies where four-digit resolution was not available for a given DRB1 or DQB1 allele in every study we have used the notation DRB1*xx00, for example DRB1*0100 to indicate that DR1 subtyping was available for some but not for all samples.
A total of 38 nonoverlapping studies of HLA DRB1-DQB1 haplotypes and T1D were analyzed from 30 countries from the following geographic regions: Sub-Saharan Africa (2 studies), North Africa (2 studies), Europe (18 studies), Middle East (3 studies), North East Asia (4 studies), South East Asia (5 studies), Oceania (1 study), and three admixed populations (Table 1). An additional seven studies supplied data only for analysis of the DR4 subsets of DRB1*0400 DQB1*0302 haplotypes. For the genotype analyses, a total of 15 populations from 14 countries were studied. In addition, for the individual genotype analyses, combined European data from Koeleman et al. (17) were studied.
| Regiona | Country/region or ethnicity | Referenceb | Studyc | Patient chromosomed | Control chromosomed | No. of haplotypes DR-DQe | No. of genotypes DR-DQe |
|---|---|---|---|---|---|---|---|
| A. All haplotypes | |||||||
| SSA | Zimbabwe | (51) | C/C | 76 | 130 | 8 | |
| SSA | Ethiopia | (52) | C/C | 78 | 78 | 5 | |
| NAF | Morocco/Souss region | (53)* | C/C | 250 | 186 | 18 | |
| NAF | Algeria/Oran region | (54) | C/C | 100 | 92 | 8 | |
| EUR | Spain/Cantabria | (55) | C/C | 164 | 206 | 19 | |
| EUR | Spain/Basque | (43) | F1 | 122 | 122 | 11 | |
| EUR | Italy/Sardinia | (16)* | C/C | 2104 | 3834 | 33 | 70 |
| EUR | Italy1/Lazio region | (56)* | C/C | 268 | 256 | 26 | 6 |
| EUR | Italy2/Northern | (43) | C/C | 252 | 212 | 20 | |
| EUR | USA/HBDI collection | (13)* | F2 | 566 | 399 | 25 | 24 |
| EUR | UK1/Bart’s-Oxford | (57)* | F1 | 1506 | 1506 | 19 | 31 |
| EUR | UK2/ BDA collection | (17) | F2 T/NT | 541 | 478 | 21 | |
| EUR | Germany | * | C/Cf | 358 | 1866 | 21 | |
| EUR | Netherlands | (17) | F1 T/NT | 352 | 337 | 15 | |
| EUR | Denmark | (43) | F1 | 104 | 104 | 7 | |
| EUR | Norway | (17) | F1 T/NT | 749 | 739 | 17 | |
| EUR | Sweden | (43) | C/C | 1257 | 1002 | 23 | 58 |
| EUR | Finland1 | (58)* | F1 | 1244 | 1244 | 18 | 63 |
| EUR | Finland2 | (43) | C/C | 126 | 126 | 13 | |
| EUR | Poland | (43) | C/C | 328 | 170 | 15 | |
| EUR | Hungary | (59)* | C/C | 298 | 354 | 19 | 17 |
| EUR | Slovenia | (60)* | C/C | 342 | 234 | 19 | 9 |
| MEA | Turkey | (61)* | C/C | 356 | 496 | 31 | 10 |
| MEA | Israel/Jewish | (62)* | C/C | 144 | 264 | 23 | |
| MEA | Israel/Arab | (62)* | C/C | 74 | 218 | 20 | |
| NEA | Japan1 | (11)* | C/C | 240 | 576 | 22 | 10 |
| NEA | Japan 2 | (23) | C/C | 264 | 314 | 21 | 9 |
| NEA | Japan 3 | (63) | C/C | 160 | 380 | 20 | |
| NEA | Korea | (64) | C/C | 316 | 280 | 23 | |
| SEA | Taiwan1 | (65) | C/C | 176 | 210 | 7 | |
| SEA | Taiwan2 | (66) | C/C | 92 | 196 | 18 | |
| SEA | Taiwan3 | (67)* | C/C | 196 | 408 | 18 | |
| SEA | Hong Kong/ S. Chinese | (68)* | C/C | 152 | 500 | 22 | 4 |
| SEA | Singapore/ S. Chinese | (69)* | C/C | 146 | 160 | 16 | 3 |
| OCE | Philippines | (70)* | C/C | 180 | 382 | 19 | 6 |
| ADM | Mexico (Mestizo) | (43) | C/C | 106 | 294 | 16 | |
| ADM | Puerto Rico/ Hispanic | (71)* | C/C | 204 | 192 | 22 | |
| ADM | USA Mexican American | (72)* | C/C | 168 | 136 | 20 | 2 |
| Country | Referenceb | Studyc | Patient chromosomed | Control chromosomed | No. of haplotypes DR-DQe | |
|---|---|---|---|---|---|---|
| B. DR4 haplotypes only | ||||||
| EUR | France | (12)* | C/C | 216 | 51 | 5 |
| EUR | Belgium | (73) | C/C | 171 | 49 | 5 |
| EUR | Germany and Belgium | (74) | T/NT | 165 | 55 | 5 |
| EUR | Germany | (74) | C/C | 490 | 340 | 7 |
| EUR | Estonia | (75) | C/C | 69 | 49 | 4 |
| EUR | Latvia | (75) | C/C | 85 | 14 | 4 |
| EUR | Russia | (75) | C/C | 159 | 85 | 4 |
- a Denotes geographic region following the anthropology/human diversity analyses of the 13th IHW (45, 76): SSA, Sub-Saharan Africa; EUR, Europe; MEA, Middle East; NEA, North East Asia; SEA, South East Asia; OCE, Oceania; ADM, admixed.
- b * denotes that the data as used were obtained by personal communication, the German data were supplied by BOB, the data from the 13th International Histocompatbility Workshop (43) are available from dbMHC at www.ncbi.nlm.nih.gov/mhc.
- c Study type: C/C, case/control; F1, family-based data ascertained for the presence of at least one affected child; F2, family-based data ascertained for the presence of at least two affected sibs; T/NT denotes that the data analyzed was transmitted (T) vs nottransmitted (NT) from parents heterozygous for the DR-DQ haplotypes.
- d The total number of chromosomes from patients and controls, respectively, is listed. For family data, the control numbers represent the total number of Affected Family BAsed Controls (AFBACs) (parental nontransmitted haplotypes (77, 78) except for T/NT data where it is the number of NT haplotypes.
- e Number of different HLA DRB1-DQB1 haplotypes or genotypes, only those haplotypes or genotypes where the patient and control counts were at least four were used in the analyses. In addition, haplotypes or genotypes observed only in one study were not included.
- f The data for the German study supplied by BOB did not contain DR4 subtypes for controls; these were estimated using the German control data from Donner et al. (74).
Sample sizes and the numbers of HLA DRB1-DQB1 haplotypes and genotypes studied from each population are listed in Table 1. Each study had on average 362 T1D patients (range, 2n= 74–2104, where n is the number of chromosomes) and 492 controls (range, 2n= 78–3834). For inclusion in the study, high-resolution typing was preferred and was required for DR4 subtypes. Some relatively older data sets are included especially if they were from a region of particular interest, for example Sub-Saharan Africa. For description of the individual studies, see the papers referenced in Table 1, although much of the data analyzed were supplied by the authors as the complete haplotype and/or genotype data were an extension of that listed in the papers. For a haplotype or genotype to be included in our analyses, the combined patient and control count had to be four or more.
For family data, the control numbers represent the total number of Affected Family BAsed Controls (AFBACs) (parental nontransmitted haplotypes) except for transmitted/nontransmitted data where it is the number of nontransmitted haplotypes (Table 1).
For countries with multiple studies (Table 1), a combined P/C ratio is listed for that country, resulting in a total of 31 data sets from 30 countries from the 38 studies. Sardinia was considered distinct from mainland Italy because of known differences between these regions. Where more than one study was available for a country, the P/C ratio was the average of the various studies regardless of the sample size in each study. Where the genotyping resolution of the various studies was not consistent (e.g. DRB1*0100 or DRB1*0101), we have used the lowest resolution so that the specificity of many alleles is limited to two-digit resolution. However, in individual studies, analyses can of course be performed at the four-digit level when this information is available.
Methods
The P/C ratio
To compare populations, we introduce a new measure, the P/C ratio, under the assumption that the HLA DR-DQ genes are directly involved in disease:
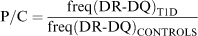
where freq(DR-DQ) denotes the frequency of the haplotype or genotype under consideration (or allele or amino acid as appropriate). This equation can also be appropriately modified for any other genetic system where a disease-predisposing locus has been identified and may include one amino acid site, or gene, or multiple genes in a region.
HLA DR-DQ genotype frequencies in the general (control) population are denoted by f(AiAj) and absolute penetrance values by wij (i≤j), where i, j= 1,2, …, k (Table 2). (The DR-DQ locus is considered as one multi-locus site denoted by locus A.) Note that these wij values represent penetrance values averaged over the effects of other T1D loci, both HLA and non-HLA. In the simplest case, the HLA DR-DQ effects would be independent of these other effects and would affect the penetrance values equally by a constant multiplicative factor. Haplotype DR-DQ frequencies in the control population are denoted by f(Ai), and their marginal (averaged over genotypes) absolute penetrance values by wi·. The genotype and haplotype frequencies in the general (control) and patient populations are given in Table 2, along with the P/C ratios. The analogy between disease models and selection models without recombination has previously been noted (6) and is illustrated here.
| Genotypes | Haplotypes | |
|---|---|---|
| AiAj | Ai | |
| Controls | f(AiAj) | f(Ai) |
| Patient population | wij f(AiAj)/T | wi · f(Ai)/T |
| P/C | wij/T | wi ·/T |
- P/C, patient/control.T=Σwij f(AiAj) is the disease prevalence.
The P/C ratios give a maximum likelihood estimate of the relative penetrance values for each genotype (fij=wij/T) and haplotype (fi.=wi·/T), i.e. the absolute penetrance values normalized by the disease prevalence in that population (proof not shown) (Table 2). Unfortunately, the P/C ratios per se cannot be directly compared across populations and particularly across ethnic groups. However, within a population, the ratio for two different genotypes or haplotypes of their P/C ratios is a function only of the absolute penetrance values, allowing comparisons across studies and ethnic groups, i.e.
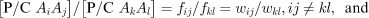
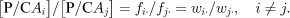
The population prevalence parameter T cancels from the relative penetrance estimates in this case, allowing direct comparisons between risk estimates from populations with different prevalences.
Hence, the ratio of two P/C ratios estimates the ratios of the absolute penetrance values for the two haplotypes or genotypes under consideration and is equivalent to the odds ratio for the comparison of these two haplotypes or genotypes. Koeleman et al. (17) applied a similar approach in that they used a consistent base of the DRB1*0101 DQB1*0501 (DR1) haplotype in their analyses of DRB1 DQA1 DQB1 haplotype effects in five European populations. The absolute penetrance values may vary between populations because they include the averaged effects of non-HLA genes, environmental factors, and other HLA genes. As mentioned above, in the simplest case, the ratio of the absolute penetrance values for the DR-DQ genotypes and haplotypes would be the same across all studies; and this is our null hypothesis.
Ranking
The rank of a haplotype or genotype was assigned 1 = most predisposing = highest P/C ratio, so that a larger ranking number corresponds to a more protective haplotype or genotype. If two haplotypes or genotypes had the exact P/C ratio, they were given the same rank. The relative ranks of haplotypes and genotypes within a study can be used in across study comparisons.
Pairwise tests
Within each population where both haplotypes were present [e.g. DR3 (DRB1*0301-DQB1*0201) and DRB1*0401 DQB1*0302], the haplotype counts in patients and controls were compared in a 2 × 2 contingency table. The P value from the corresponding Pearson’s chi-square was computed. Standard linear regression methods were carried out to test for correlation and significance level between pairs of populations or between populations and the mean P/C ratio for a given haplotype using S-plus 6.0 (Insightful Corp, Seattle, WA).
Results
P/C ratios and relative ranks of DR-DQ haplotypes
In Table 3, the P/C ratios are listed by country, with the DRB1-DQB1 haplotypes listed in order of their mean P/C ratios, from most predisposing DRB1*0405 DQB1*0302 to most protective DRB1*1500 DQB1*0602. The average rank is also considered, and there is good agreement between ordering by the mean P/C ratios and the average rank. The same general pattern was also seen using all studies (data not shown). We must note that neither measure is perfect; P/C ratios will vary across populations and particularly across ethnic regions as they are a function of disease prevalence, and most haplotypes do not occur in all studies; a factor that also makes the average ranking not a perfect measure. However, combined with the pairwise analyses, the ordering from most predisposing to least predisposing T1D haplotypes is in general consistent.
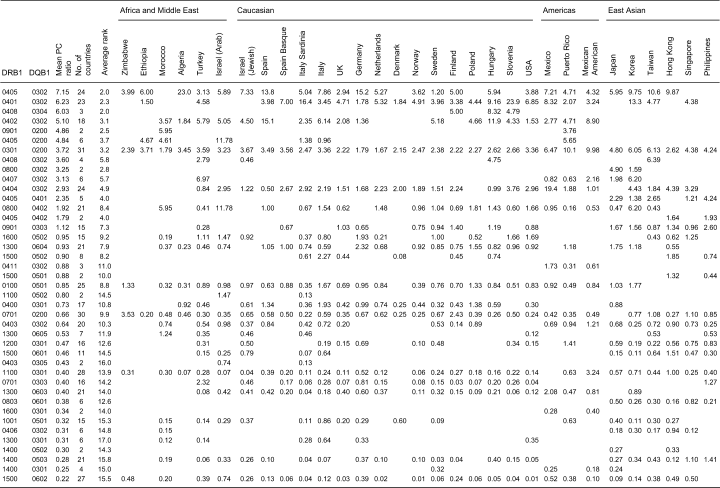 |
For all pairs of populations that had at least five haplotypes in common, we assessed how correlated were the P/C ratios. Over 255 such comparisons, we found an average Pearson’s correlation coefficient of 0.804 (R2= 0.65) and an average statistical significance of P < 0.0002.
In addition, for the 21 haplotypes present in at least 11 countries, we explored the correlation between the observed P/C ratio in individual populations and the mean PC ratios over all populations. By linear regression, we computed the square of the correlation coefficient and the P value for the four ethnic groupings (Figure 1). The overall correlation between mean P/C ratio and individual population P/C ratio yielded an R2= 0.49 and P < 5 × 10−66. Interestingly, the best correlation between the overall mean P/C ratio and the individual ones for populations was seen in the East Asian countries. The worst fit was seen in the three countries from the Americas.
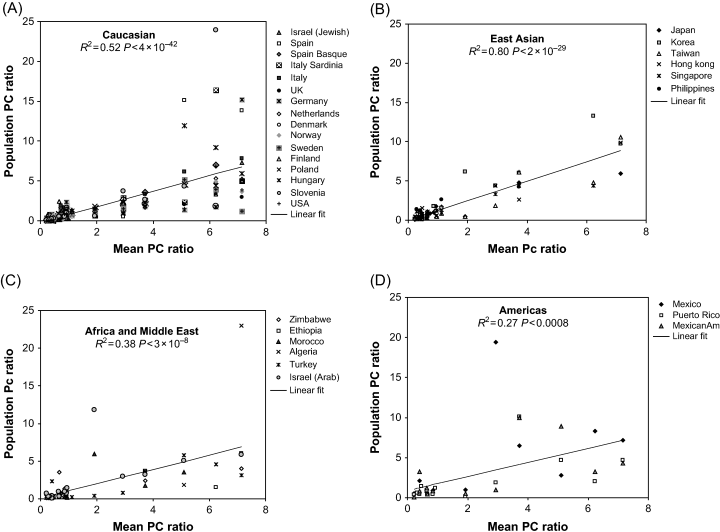
Correlation between patient/control (P/C) ratios in individual populations and the mean P/C ratio for the 21 haplotypes represented in most countries. The mean PC ratio is over all populations, the linear fits shown are for each of the four ethnic groupings (A) Caucasians (B) East Asian (C) Africa and Middle East (D) Americas.
In this study, we use the mean P/C ratios to give a relative initial ordering of RPEs. Statistical differences can be inferred when specific combinations are compared within and across studies as shown below, using either 2 × 2 contingency table comparisons or the relative rank orders in each study.
General trends are as previously well documented. There is obviously a continuous gradient from highly predisposing to very protective haplotypes. However, we will continue the tradition, which is reasonable, of often referring to common haplotypes in categories of highly predisposing through to predisposing [the DRB1*0405, 0401, and 0402 haplotypes with DQB1*0302, the DR3 haplotype (DRB1*0301 DQB1*0201) and the DRB1*0404 DQB1*0302 haplotype], then a ‘neutral’ (intermediate) category including DRB1*0800 DQB1*0402, DRB1*0901 DQB1*0303 (DR9), and DRB1*0100 DQB1*0501 (DR1), followed by the protective to very protective category culminating in the most protective haplotypes such as DRB1*1400 DQB1*0503 and DRB1*1500 DQB1*0602.
Meta-analyses comparing relative ranks of pairs of haplotypes can be carried out from this table, for example, comparing DR3 and DR9 haplotypes, the P/C ratio for DR3 is greater than that for DR9 in all 15 data sets where they both occur (Table 3), and in fact considerably larger in all cases. Similarly, as is well known, DRB1*0401 DQB1*0302 is strongly and consistently more predisposing than the DRB1*0401 DQB1*0301 haplotype; this original observation led of course to the Asp at DQB1 position 57 protective vs non-Asp predisposing hypothesis. For these and other comparisons, no distinct ethnic group differences are seen. Other comparisons will be considered below.
Pairwise tests including single Amino acid site comparisons
In combination with the meta-analysis across populations (Table 3), we performed pairwise tests for a number of haplotype combinations (Table 4), in particular those that were present in a number of countries and at high frequency in some countries. We subdivided the haplotypes into sets of interest for studying their relative predisposition (Table 4A,B, DR3, and DRB1*0401, 0402, 0405, and 0404 with DQB1*0302), neutral (Table 4C,D), and protective (Table 4E,F). The number of populations where both haplotypes tested were present is shown, as is the percent where one or the other had a higher P/C ratio and the percent (over the total number of populations with this haplotype) where this difference was statistically significant with P < 0.05. When significant pairwise comparisons involve only one allele of DRB1 or DQB1 but not both being different, then the amino acids will be mentioned when these are of interest, i.e. if only one or a few amino acids are involved.
| A | ||||||
|---|---|---|---|---|---|---|
| 0301 0201 vs | No. of studies | % DR4-0302 more predisposing than DR3 | P < 0.05 | % DR4-0302 less predisposing than DR3 | P < 0.05 | DR4x/DR3a |
| 0401 0302 | 26 | 76.9 | 46.5% | 23.1 | 0.0% | 2.11 |
| 0402 0302 | 19 | 63.2 | 5.3% | 36.8 | 0.0% | 1.56 |
| 0405 0302 | 26 | 80.8 | 11.5% | 19.2 | 0.0% | 2.32 |
| 0404 0302 | 28 | 25.0 | 0.0% | 75.0 | 17.9% | 0.78 |
| B | ||||||
|---|---|---|---|---|---|---|
| 0401 0302 vs | No. of studies | % 0401 more predisposing | P < 0.05 | % 0401 less predisposing | P < 0.05 | 0401/DRB1*0402/4/5a |
| 0402 0302 | 23 | 47.8 | 0.0% | 52.2 | 0.0% | 1.22 |
| 0405 0302 | 24 | 45.8 | 0.0% | 54.2 | 0.0% | 1.09 |
| 0404 0302 | 33 | 90.9 | 30.2% | 9.1 | 0.0% | 3.20 |
| C | ||||||
|---|---|---|---|---|---|---|
| 09-0303 vs | No. of studies | % 09-0303 more predisposing | P < 0.05 | % 09-0303 less predisposing | P < 0.05 | 09-0303/DR1 |
| 0100 0501 | 17 | 52.9 | 11.8% | 47.1 | 0.0% | 1.35 |
| D | ||||||
|---|---|---|---|---|---|---|
| 08-0402 vs | No. of studies | % 08-0402 more predisposing | P < 0.05 | % 08-0402 less predisposing | P < 0.05 | 08-0402/DR1 or DR9 |
| 0100 0501 | 28 | 64.3 | 10.7% | 35.7 | 0.0% | 2.15 |
| 0900 0303 | 16 | 50.0 | 0.0% | 50.0 | 0.0% | 1.62 |
| E | ||||||
|---|---|---|---|---|---|---|
| 15-0602 vs | No. of studies | % 15-0602 more protective | P < 0.05 | % 15-0602 less protective | P < 0.05 | DRx/(15_0602)b |
| 0701 0303 | 18 | 88.9 | 16.7% | 11.1 | 0.0% | 3.85 |
| 1100 0301 | 32 | 81.3 | 25.0% | 18.7 | 0.0% | 4.54 |
| 1300 0603 | 24 | 79.2 | 33.3% | 20.8 | 0.0% | 6.12 |
| 1400 0503 | 24 | 62.5 | 4.2% | 37.5 | 4.2% | 2.21 |
| F | ||||||
|---|---|---|---|---|---|---|
| 14-0503 vs | No. of studies | % 14-0503 more protective | P < 0.05 | % 14-0503 less protective | P < 0.05 | DRx/(14-0503)b |
| 0701 0303 | 15 | 66.7 | 6.7% | 33.3 | 0.0% | 4.88 |
| 1100 0301 | 25 | 72.0 | 16.0% | 28.0 | 0.0% | 2.50 |
| 1300 0603 | 17 | 88.2 | 5.9% | 11.8 | 0.0% | 3.05 |
- a Mean PC(DR4)/mean PC(DR3) or mean PC(0401-0302)/mean PC(040x-0302).
- b Mean PC(other)/PC(DR15_DQ6) or mean PC(other)/PC(DR14_DQ5).
With these tests, we show that overall the DR3 haplotype is significantly less predisposing than the DRB1*0405, *0401 and *0402 with DQB1*0302 haplotypes (11.5%, 46.5%, and 5.3%, respectively, of countries with significant effects), but the DR3 haplotype is significantly more predisposing than DRB1*0404 DQB1*0302 (17.9% of countries with significant effects) (Table 4A). Within these DR4 haplotypes, the pairwise comparisons show no significant differences between DRB1*0405, *0401 and *0402 with DQB1*0302, but as expected from the DR3 results, these three DR4 haplotypes are significantly more predisposing than DRB1*0404 DQB1*0302 (Table 4B) (30.2% of countries show significant differences between DRB1*0401 and *0404).
For the intermediate-risk or neutral haplotypes (Table 4C,D), DRB1*0800 DQB1*0402 and DRB1*0901 DQB1*0303 (DR9) while not significantly different from each other are both significantly slightly more predisposing than DRB1*0100 DQB1*0501 (DR1) (10.7% and 11.8% of countries, respectively, show significant differences).
For protective haplotypes (Table 4E,F), DRB1*1500 DQB1*0602 is slightly more protective than DRB1*1400 DQB1*0503 (in 62.5% vs 37.5% of populations, but significant effects are divided equally between 4.2% of countries), and both these haplotypes are significantly more protective than DRB1*1100 DQB1*0301, DRB1*0701 DQB1*0303, and DRB1*1300 DQB1*0603.
Of interest also is further comparison of the DRB1*0400 DQB1*0302 haplotypes (4, 5, 12, 14). In accord with previous observations, the protective DRB1*0403 haplotype has a lower P/C ratio than the predisposing DRB1*0404 haplotype in 17 vs 3 populations (and these three cases involve at least one rare haplotype) and very highly significantly differences are seen in four populations. Comparisons involving DRB1*0406, 0407, and 0408 are more difficult because of their lower frequencies. DRB1*0407 is more predisposing than DRB1*0403 in six vs two populations (the latter two involve rare haplotypes) and is significantly more predisposing in three populations. A slightly higher predisposing effect for DRB1*0408 over DRB1*0404 as indicated in Table 3 has no statistical support; similarly for DRB1*0407 vs DRB1*0404, except that in the Mexican mestizos sample, there is highly significant (P < 0.00001) support for DRB1*0404 being more predisposing than DRB1*0407. In comparison of the protective DRB1*0403 and DRB1*0406 haplotypes, there is only a very slight trend (6:2) toward greater protection of the latter. Thus, our results for DRB1*0400 DQB1*0302 haplotypes can be summarized as follows, with the placement of those in parentheses lacking statistical support: 0405 = 0401 = 0402 > 0404 (0407, 0408) > 0403 (0406).
Amino acid site differences at DRB1 involving one amino acid are as follows (with the more predisposing allele listed first): *0405 vs*0408 [#57 S(Ser) vs D(Asp)], *0401 vs*0408 [#71 K(Lys) vs R(Arg)], *0407 vs*0403 [#86 G(Gly) vs V(Val)]. Note, however, that both *0405 and *0401 differ from *0402 at position 86 with again G vs V (as well as at other sites #57, 67, 70, and 71), and because these three DRB1*0400 alleles are not significantly different in risk, these results implicate combinations of sites as risk factors rather than single amino acids.
The protective effect of the DRB1*0403 allele with the predisposing DQB1*0302 allele balances out to be equivalent to the overall protective effect of the DQB1*0301 allele on DRB1*0401 haplotypes (no significant overall or individual comparisons).
For DRB1*0405 haplotypes, in three populations those with DQB1*0302 were more predisposing than those with DQB1*0201, and this difference was very highly significant in the Sardinian sample (P < 0.0000001). DQB1*0302 was more predisposing than DQB1*0401 in four of four samples, with significance in two comparisons. A number of amino acid differences separate these pairwise contrasting allele predisposition effects.
Amino acid sites
We have selected a set of residues in DRB1 and DQB1 that have either been implicated in T1D susceptibility by various authors or are in linkage disequilibrium with DQ-alpha residues that have, as DQA1 genotyping was not available in most studies (6, 12, 16–25) (Table 5). We do not claim that the residues selected can explain susceptibility to T1D, but rather we wanted to explore how consistent such combinations would be across haplotypes and ethnic boundaries.
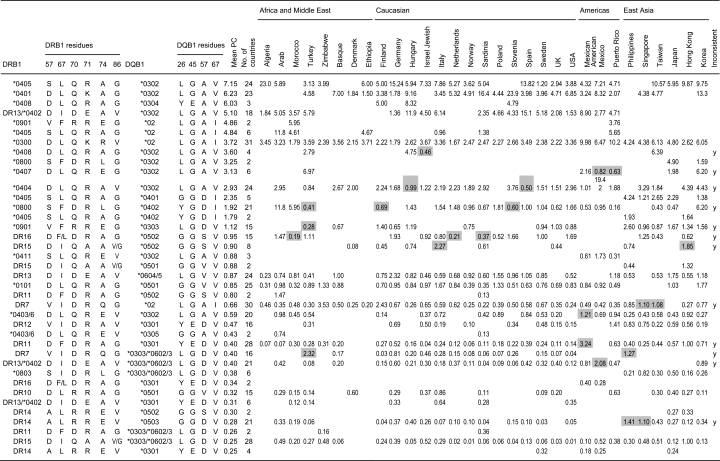 |
Where the genotype resolution available does not allow us to establish allele subtype (e.g. DQB1*0400), an ‘x’ has been used at any residue that is undetermined. If two or more alleles correspond to a given residue combination (e.g. ‘DLQRAG’ can be DR1 or DRB1*0408) but the haplotype amino acid combination does not occur in any population (e.g. DRB1*0408 DQB1*0501 was not seen in any of the populations at least once), then only the actually observed haplotype combination corresponding to a given residue sequence is presented. ‘Inconsistent’ refers to haplotypes or genotypes that have a P/C ratio > 1 in one or more populations and, also, <1 in one or more populations. The P/C ratios that are discordant with the mean ratio for that haplotype or genotype have been highlighted in gray in Table 5 and Table 7.
 |
Although these amino acids have been shown by the haplotype method to explain much of the HLA DR-DQ risk (6), nonetheless there is not a consistent hierarchical pattern of risk involving these amino acids per se. Combinations of these amino acids in specific allelic combinations appear to confer predisposing vs protective properties on the HLA DR-DQ molecules, and various combinations of amino acids can give equivalent risks.
P/C ratios and relative ranks of DR-DQ genotypes
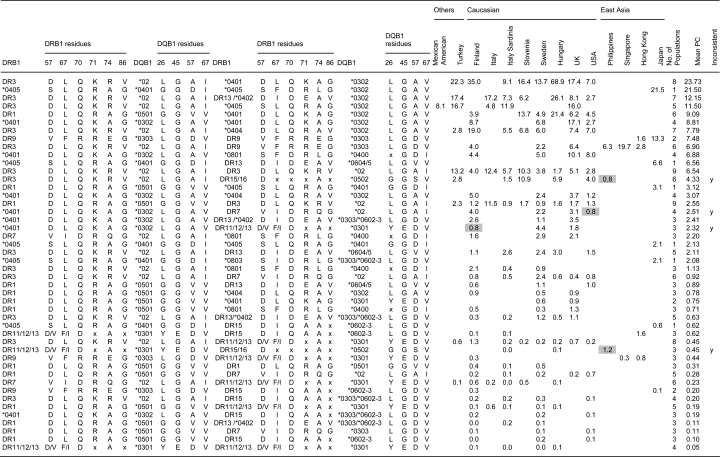
In Table 6, the P/C genotype ratios are listed by country, with the DRB1-DQB1 genotypes listed in order of their mean P/C ratios, from most predisposing to most protective. Note that with the genotype analyses, less confidence can be placed on the P/C values because in many cases, the control genotype counts were small. We have investigated various hypotheses using both the overall rankings in Table 6 and pairwise comparisons in individual populations. Table 7 includes amino acid data as well the genotype data.
| DRB1a | DQB1a | DRB1b | DQB1b | Mean PC | No. of countriesa | Finland | Hungary | Italy | Italy Sardinia | Mexican American | Slovenia | Sweden | Turkey | UK | USA | Japan | Hong Kong | Philippines | Singapore |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0300 | 0200 | 0401 | 0302 | 26.66 | 8 | 35.00 | 68.90 | 9.11 | 16.42 | 37.21 | 22.29 | 17.35 | 7.00 | ||||||
| 0405 | 0401 | 0800 | 0302 | 21.50 | 1 | 21.5 | |||||||||||||
| 0300 | 0200 | 0407 | 0302 | 21.05 | 1 | 21.05 | |||||||||||||
| 0300 | 0200 | 0402 | 0302 | 12.15 | 7 | 26.13 | 17.19 | 7.29 | 6.16 | 17.42 | 8.08 | 2.75 | |||||||
| 0300 | 0200 | 0405 | 0302 | 11.50 | 5 | 4.78 | 11.90 | 8.10 | 16.72 | 16.03 | |||||||||
| 0100 | 0501 | 0401 | 0302 | 9.59 | 6 | 3.92 | 21.38 | 13.68 | 7.89 | 6.18 | 4.50 | ||||||||
| 0404 | 0302 | 0404 | 0302 | 9.49 | 2 | 16.00 | 2.99 | ||||||||||||
| 0401 | 0302 | 0401 | 0302 | 8.81 | 4 | 8.67 | 6.82 | 17.08 | 2.67 | ||||||||||
| 0401 | 0302 | 1300 | 0604 | 8.70 | 2 | 9.00 | 8.40 | ||||||||||||
| 0300 | 0200 | 0404 | 0302 | 7.74 | 7 | 19.00 | 5.47 | 6.84 | 5.66 | 2.79 | 7.45 | 7.00 | |||||||
| 0401 | 0302 | 0800 | 0400 | 7.51 | 4 | 4.36 | 7.55 | 10.13 | 8.00 | ||||||||||
| 0900 | 0303 | 0900 | 0303 | 7.48 | 2 | 13.3 | 1.64 | ||||||||||||
| 0300 | 0200 | 0900 | 0303 | 6.90 | 6 | 4.00 | 2.19 | 6.37 | 2.82 | 6.27 | 19.7 | ||||||||
| 0405 | 0401 | 1300 | 0604 | 6.56 | 1 | 6.56 | |||||||||||||
| 0300 | 0200 | 0300 | 0200 | 6.54 | 9 | 4.00 | 1.66 | 12.42 | 5.65 | 10.26 | 3.77 | 13.24 | 5.09 | 2.80 | |||||
| 0401 | 0302 | 0900 | 0303 | 6.53 | 2 | 4.33 | 8.73 | ||||||||||||
| 0300 | 0200 | 0405 | 0401 | 5.48 | 1 | 5.48 | |||||||||||||
| 0300 | 0200 | 15/16 | 0502 | 4.33 | 6 | 5.94 | 1.50 | 10.95 | 2.79 | 4.00 | 0.84 | ||||||||
| 0404 | 0302 | 1300 | 0604 | 3.33 | 2 | 6.00 | 0.67 | ||||||||||||
| 0404 | 0302 | 0700 | 0200 | 3.27 | 2 | 6.00 | 0.55 | ||||||||||||
| 0100 | 0501 | 0405 | 0401 | 3.12 | 1 | 3.12 | |||||||||||||
| 0401 | 0302 | 0404 | 0302 | 3.07 | 4 | 5.00 | 2.37 | 3.69 | 1.20 | ||||||||||
| 0100 | 0501 | 0300 | 0200 | 2.89 | 9 | 1.16 | 1.58 | 11.46 | 0.90 | 1.71 | 3.87 | 2.32 | 1.68 | 1.29 | |||||
| 0401 | 0302 | 0700 | 0200 | 2.51 | 4 | 4.00 | 2.19 | 3.10 | 0.75 | ||||||||||
| 0401 | 0302 | 1300 | 0603 | 2.43 | 3 | 2.60 | 1.17 | 3.51 | |||||||||||
| 0401 | 0302 | 11/12/13 | 0301 | 2.32 | 3 | 0.79 | 4.38 | 1.79 | |||||||||||
| 0700 | 0200 | 0800 | 0400 | 2.20 | 3 | 1.60 | 2.92 | 2.08 | |||||||||||
| 0405 | 0401 | 0405 | 0401 | 2.13 | 1 | 2.13 | |||||||||||||
| 0300 | 0200 | 1300 | 0604 | 2.11 | 5 | 1.13 | 2.97 | 2.60 | 2.37 | 1.50 | |||||||||
| 0405 | 0401 | 0803 | 0601 | 2.08 | 1 | 2.08 | |||||||||||||
| 0405 | 0401 | 0900 | 0303 | 2.05 | 1 | 2.05 | |||||||||||||
| 0100 | 0501 | 0405 | 0302 | 1.77 | 2 | 1.06 | 2.47 | ||||||||||||
| 0300 | 0200 | 0800 | 0400 | 1.13 | 3 | 2.14 | 0.40 | 0.85 | |||||||||||
| 0100 | 0501 | 0900 | 0303 | 1.12 | 2 | 1.14 | 1.10 | ||||||||||||
| 0401 | 0301 | 0401 | 0302 | 1.10 | 2 | 0.85 | 1.34 | ||||||||||||
| 0900 | 0303 | 15/16 | 0502 | 1.04 | 1 | 1.04 | |||||||||||||
| 0300 | 0200 | 0700 | 0200 | 0.92 | 6 | 0.75 | 0.59 | 0.52 | 2.44 | 0.43 | 0.80 | ||||||||
| 0100 | 0501 | 1300 | 0604 | 0.89 | 3 | 0.57 | 1.10 | 1.00 | |||||||||||
| 11/12/13 | 0301 | 1500 | 0601 | 0.88 | 2 | 0.11 | 1.64 | ||||||||||||
| 0300 | 0200 | 0401 | 0301 | 0.83 | 2 | 0.99 | 0.68 | ||||||||||||
| 11/12/13 | 0301 | 1500 | 0501 | 0.78 | 1 | 0.78 | |||||||||||||
| 0100 | 0501 | 0404 | 0302 | 0.78 | 3 | 0.89 | 0.55 | 0.89 | |||||||||||
| 0100 | 0501 | 0401 | 0301 | 0.75 | 3 | 0.58 | 0.91 | ||||||||||||
| 0100 | 0501 | 0800 | 0400 | 0.71 | 3 | 0.52 | 0.31 | 1.30 | |||||||||||
| 0300 | 0200 | 1400 | 0503 | 0.65 | 2 | 1.19 | 0.11 | ||||||||||||
| 0300 | 0200 | 1300 | 0603 | 0.63 | 5 | 0.25 | 0.48 | 0.15 | 1.22 | 1.06 | |||||||||
| 0405 | 0401 | 1500 | 0601 | 0.62 | 1 | 0.62 | |||||||||||||
| 0404 | 0302 | 1500 | 0602 | 0.52 | 2 | 0.86 | 0.18 | ||||||||||||
| 0404 | 0302 | 1300 | 0603 | 0.50 | 2 | 0.80 | 0.21 | ||||||||||||
| 11/12/13 | 0301 | 15/16 | 0502 | 0.47 | 3 | 0.08 | 0.09 | 1.22 | |||||||||||
| 0300 | 0200 | 11/12/13 | 0301 | 0.46 | 8 | 1.33 | 0.24 | 0.29 | 0.17 | 0.24 | 0.56 | 0.67 | 0.20 | ||||||
| 0900 | 0303 | 11/12/13 | 0301 | 0.44 | 3 | 0.29 | 0.76 | 0.27 | |||||||||||
| 0100 | 0501 | 0100 | 0501 | 0.31 | 3 | 0.35 | 0.09 | 0.49 | |||||||||||
| 1300 | 0603 | 1300 | 0603 | 0.29 | 2 | 0.10 | 0.49 | ||||||||||||
| 0100 | 0501 | 0700 | 0200 | 0.28 | 5 | 0.23 | 0.12 | 0.18 | 0.18 | 0.67 | |||||||||
| 0100 | 0501 | 15/16 | 0502 | 0.27 | 2 | 0.30 | 0.24 | ||||||||||||
| 0800 | 0400 | 1500 | 0602 | 0.26 | 2 | 0.02 | 0.49 | ||||||||||||
| 0700 | 0200 | 11/12/13 | 0301 | 0.23 | 6 | 0.57 | 0.07 | 0.16 | 0.03 | 0.46 | 0.12 | ||||||||
| 0900 | 0303 | 1500 | 0602 | 0.23 | 2 | 0.33 | 0.12 | ||||||||||||
| 0300 | 0200 | 1500 | 0602 | 0.21 | 4 | 0.18 | 0.26 | 0.32 | 0.08 | ||||||||||
| 15/16 | 0502 | 15/16 | 0502 | 0.21 | 2 | 0.15 | 0.26 | ||||||||||||
| 0401 | 0302 | 1500 | 0602 | 0.20 | 3 | 0.25 | 0.28 | 0.08 | |||||||||||
| 0800 | 0400 | 1300 | 0604 | 0.20 | 2 | 0.25 | 0.15 | ||||||||||||
| 0100 | 0501 | 11/12/13 | 0301 | 0.19 | 5 | 0.11 | 0.07 | 0.64 | 0.08 | 0.08 | |||||||||
| 11/12/13 | 0301 | 1300 | 0604 | 0.17 | 2 | 0.15 | 0.18 | ||||||||||||
| 0100 | 0501 | 1300 | 0603 | 0.11 | 3 | 0.03 | 0.15 | 0.15 | |||||||||||
| 0100 | 0501 | 0700 | 0303 | 0.11 | 3 | 0.07 | 0.15 | 0.10 | |||||||||||
| 11/12/13 | 0301 | 1300 | 0603 | 0.10 | 2 | 0.08 | 0.11 | ||||||||||||
| 0100 | 0501 | 1500 | 0602 | 0.10 | 3 | 0.03 | 0.18 | 0.08 | |||||||||||
| 0900 | 0303 | 1500 | 0601 | 0.07 | 1 | 0.07 | |||||||||||||
| 1500 | 0602 | 1500 | 0602 | 0.06 | 2 | 0.03 | 0.10 | ||||||||||||
| 1300 | 0603 | 1500 | 0602 | 0.06 | 2 | 0.03 | 0.09 | ||||||||||||
| 0700 | 0200 | 1500 | 0602 | 0.06 | 2 | 0.05 | 0.06 | ||||||||||||
| 11/12/13 | 0301 | 11/12/13 | 0301 | 0.05 | 4 | 0.10 | 0.05 | 0.02 | 0.05 | ||||||||||
- a All the Asian genotypes are shown. Only those Caucasian genotypes seen in at least two populations are shown.
The increased risk of various DR3/DR4 combinations
The well-known increased risk for DR3/DR4 heterozygotes over their respective homozygotes for highly predisposing DR4 subtypes (DRB1*0401, 0402, and 0405 with DQB1*0302) is seen in this data set both in overall analyses (Table 6) and in individual population comparisons. The small numbers in some categories for each of these DR3/DR4 heterozygous combinations does not allow any distinction in risk to be studied for the DR4 subtypes.
Of interest is that the DR3/DRB1*0404 DQB1*0302 heterozygote has a slightly higher risk than the DR3 homozygote, although this DR4 subtype has significantly lower overall haplotype risk than DR3. However, no statistical weight can be given to this observation with these data.
Is DRB1*0401 DQB1*0302 vs DQB1*0301 increased in DR3 relative to DR1 heterozygotes?
Noble et al. (29) observed a trend for an increased frequency of DRB1*0401 DQB1*0301 vs DQB1*0302 in DR1 vs DR3 heterozygotes, or equivalently an increased frequency of DRB1*0401 DQB1*0302 vs DQB1*0301 in DR3 vs DR1 heterozygotes. This trend continues to be observed in the current USA/HBDI collection as well as in three other populations with sufficient sample size. The odds ratios for DR3 heterozygotes with DRB1*0401 DQB1*0302 vs DRB1*0401 DQB1*0301 and similarly for DR1 heterozygotes are as follows: UK1 25.5 (DR3s), 6.8 (DR1s), Finland1 15.0, 3.9, and 17.6 and 4.3 for a combined European sample [Italy/Sardinia, UK2, Norway, The Netherlands, and a subset of the USA/HBDI collection (17)]. That is, DRB1*0401 DQB1*0302 is relatively more frequent than DRB1*0401 DQB1*0301 in DR3 heterozygotes than in DR1 heterozygotes.
Do DR3/DR9s have increased risk over DR3/DR3s?
The predisposing DR3 haplotype is relatively rare in Asian populations, while the neutral DR9 haplotype is more frequent in the Asian than in the Caucasian populations. The genotype DR3/DR9 can be relatively common in Asian patients, raising the possibility that DR3/DR9s have increased risk over DR3/DR3s and/or DR9/DR9s. From the overall rankings and a few instances where individual population comparisons can be made, there is no evidence for differential risk. This reflects mostly lack of statistical power rather than confidence in measuring the true effects.
Discussion
Our results show the strength of meta-analysis combined with population-specific comparisons. While previous studies have suggested trends of differences in predisposing effects between the DRB1*0401, *0402, and *0405 with DQB1*0302 haplotypes (12, 14), our analyses show that there is no statistical support for such differences, including across ethnic groups.
The comparison of, for example, DR3 and DR9 effects show the importance of looking at the P/C ratios to determine the relative predisposing effects and their strengths. Other measures such as the relative risk where a haplotype is compared with the ‘other’ category, and P values from tests of heterogeneity, suffer from variation across populations and ethnic groups of multiple predisposing effects of the other haplotypes and sample size issues. Our results show that the relative categorization worldwide of DR3 as predisposing and DR9 as neutral is correct; the common occurrence of DR9 in Asian T1D patients, and in particular DR3/DR9, is because of the high frequency of the DR9 haplotype relative to the predisposing category of haplotypes that are relatively common in the Caucasian populations. DR3 is much less frequent in Asian populations; yet, it still maintains its predisposing effect when it occurs.
With regard to the P/C ratio, a weakness that must be mentioned is the potentially large variance because of the fact that it is a ratio of two variables. Further, of course the accuracy of the estimate of the P/C ratio is very dependent on sample size. However, its strength is that its mean value directly reflects the relative penetrance values of each haplotype.
It is not possible to list a set of amino acids that give a linear relationship with T1D risk, this is readily appreciated given the number of amino acid differences between the highly predisposing DRB1*0400 DQB1*0302 haplotypes and the DR3 haplotype. With the hierarchy of risk when all DRB1*0400 DQB1*0302 haplotypes are considered, there are some single amino acid changes that correlate with significantly different risks among these haplotypes, but even here in some cases, combinations of amino acids are needed to explain effects.
The problem of sample size is immediately obvious in comparison of genotype effects both within and across populations (Table 6). Combined with the lack of genotype data for many studies and that when it is available very few genotype combinations have reasonable numbers of observed in patients and particularly in controls, we are unable to make many meaningful comparisons. The well-known increased risk for DR3/DR4s with predisposing DR4 subtypes is obvious. Furthermore, significant genotype-specific effects (DR3 vs DR1) are seen for DQB1*0302 vs DQB1*0301 on DRB1*0401 haplotypes.
Further studies require high-resolution HLA DR-DQ typing of extremely large samples combined with cross ethnic studies to obtain more accurate predictors of T1D risk at the genotype level. Current studies by the Type 1 Diabetes Genetic Consortium will aid in this respect.
Acknowledgments
We thank Andrea Cabello, Kitty Deng, Sheila Rajashekara, Shweta Patel, Howard Ching, and Neha Prakash for data entry. Research supported by NIH grants GM56688 and GM35326 (GT, AMV, MNG), R24 CA84497 (GT, AP, AS, JSD, SC-Z), NIH DK61722 (AMV, JAN, HAE), NIH DK46626 (JAN, HAE), Hungarian Scientific Research Fund (OTKA) No. T046923 (RH), Juvenile Diabetes Research Foundation, European Foundation for the Study of Diabetes, NOVO NORDISK (RH, JI), Wellcome Trust and Diabetes UK (APL, PJB, KMG), Diabetes Incidence in Sweden Study Group and Swedish Childhood Diabetes Study Group (ÅL, IK, CBS), the Swedish Diabetes Foundation and Barndiabetes Fonden (IK), Swedish Medical Research Council Grant (CBS, ÅL), NIH grants DK53004 and DK26190 (ÅL), the Norwegian Research Council (166515) (KSR), FIRB2001: RBNE01C5S2_004 (AP, RB), Dutch Diabetes Research Foundation (97.137, 2001.10.004), the Netherlands Organisation for Health Research and Development (ZonMW) and the Juvenile Diabetes Research Foundation International (JDRF) (BPCK, BOR), the Turkish Scientific and Technical Research Council (TUBITAK SBAG-1529) (GS-D), Deutsche Forschungsgemeinschaft (DFG, SFB 518, GRK 1041) (BOB), the Polish Ministry of Education and Science grant 2P05E05129 (WM), Grant-in-aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology, Japan (HI), Israeli Ministry of Health Chief Scientist Office Grant 3228 (SI), MBRS and Extramural Associates Sponsored Research (AS-C, TFL), NIH HD33674 (AS-C), 2RO1 HD37800, 4R33 DK069878-03, 4R33 HD050196 (J-XS), Cedars-Sinai Board of Governors’ Chair in Medical Genetics (JIR), grant 97/1119 from the Fondo de Investigaciones Sanitarias of the Ministry of Health of Spain and a grant from the ‘Marqués de Valdecilla’ Foundation (FL-C), Fondo de Investigacion Sanitaria (FIS) del Instituto de Salud Carlos III, Spain (grants G03/212, C03/08, and PI05/2291) (LC).




