OsRAD21-3, an orthologue of yeast RAD21, is required for pollen development in Oryza sativa
Summary
In contrast to animals, in which products of meiosis differentiate directly into sperm, flowering plants employ a specific mechanism to give rise to functional sperm cells, the specifics of which remain largely unknown. A previous study revealed that, compared to yeast and vertebrates, which have two proteins (Rad21 and its meiosis-specific variant Rec8) that play a vital role in sister chromatid cohesion and segregation for mitosis and meiosis, respectively, the rice genome encodes four Rad21/Rec8 proteins (OsRad21s). In this paper, phylogenetic and immunostaining analyses reveal that OsRad21-3 is an orthologue of yeast Rad21. OsRAD21-3 transcript and protein accumulated preferentially in flowers, with low levels in vegetative tissues. In flowers, they persisted from the stamen and carpel primordia stages until the mature pollen stage. OsRAD21-3-deficient RNAi lines showed arrested pollen mitosis, aberrant pollen chromosome segregation and aborted pollen grains, which led to disrupted pollen viability. However, male meiosis in these RNAi lines did not appear to be severely disrupted, which suggests that the main involvement of OsRAD21-3 is in post-meiotic pollen development by affecting pollen mitosis. Furthermore, of the four OsRAD21 genes in the rice genome, only OsRAD21-3 was expressed in pollen grains. Given that the mechanism involving generation of sperm cells differs between flowering plants and metozoans, this study shows, in part, why flowering plants of rice and Arabidopsis have four Rad21/Rec8 proteins, as compared with two in yeast and metozoans, and gives some clues to the functional differentiation of Rad21/Rec8 proteins during evolution.
Introduction
In flowering plants, the highly specialized haploid male gametophyte (pollen) that is generated from diploid pollen mother cells (PMCs) in anthers is a key regulator of sexual reproduction and contributes to the genetic diversity of a population. The formation of pollen involves two principal phases: microsporogenesis and microgametogenesis. During microsporogenesis, PMCs are differentiated from archesporial cells, and thereafter undergo meiosis to generate haploid microspores. During microgametogenesis, the microspore released from tetrads quickly increases in size and then undergoes asymmetric mitosis (pollen mitosis I, PMI) to yield a large vegetative cell and a small generative cell. The large vegetative cell exits the cell cycle, and contains a dispersed nucleus and most of the cytoplasm from the microspore, whereas the diminutive generative cell has a condensed nucleus and a small amount of cytoplasm, and undergoes a second mitosis (pollen mitosis II, PMII), generating two sperm cells (McCormick, 2004; Twell, 2002; Wilson and Yang, 2004). In some plants, the second mitosis occurs in the growing pollen tube, whereas in other species, such as grasses and crucifers, it is completed before anthesis (Ma, 2005; McCormick, 2004). In contrast to the formation of germ cells in animals (Wilson and Yang, 2004), the two cycles of mitosis at the post-meiotic stage are specific to flowering plants.
Recent efforts to decipher molecular events underlying male meiosis have extended our understanding of pollen development, in particular genetic studies, which have identified a number of meiotic mutants in Arabidopsis thaliana, Zea mays and Oryza sativa (Ma, 2005). Furthermore, forward and reverse genetic studies have functionally identified some key genes involved in male meiosis, including those involved in homologue pairing, recombination, repair and sister chromatid cohesion in these plants (Ma, 2005). In contrast, although some mutants involved in male mitosis have been identified in Arabidopsis (Chen and McCormick, 1996; Durbarry et al., 2005; Iwakawa et al., 2006; Lalanne et al., 2004; McCormick, 2004; Park et al., 1998; Twell, 2002), our knowledge of the mechanism underlying male mitosis is still limited.
Meiosis and mitosis involve a series of events, including sister chromatid cohesion, that are coordinated in time to guarantee faithful segregation of chromosomes. The cohesion binds newly replicated sister chromatids from S phase until the metaphase–anaphase transition, and is mediated by a phylogenetically conserved multi-protein cohesin complex (Lee and Orr-Weaver, 2001; Nasmyth, 2001). Mitotic cohesion in Schizosaccharomyces pombe and Saccharomyces cerevisiae involves at least Rad21 (also called Scc1 or Mcd1), Scc3, Smc1 and Smc3, and Rad21 is a vital subunit of the complex (Guacci et al., 1997; Michaelis et al., 1997). At the metaphase–anaphase transition, proteolytic cleavage of Rad21 mediated by separase triggers dissociation of cohesion proteins from chromosomes. The cleavage is necessary and sufficient for sister separation (Tomonaga et al., 2000; Uhlmann et al., 1999, 2000). In meiotic cohesin of both fission and budding yeast, Rad21 is replaced by its meiotic variant Rec8; this replacement is essential for differential release of the chromosome arm and centromeric cohesion in meiosis I and II (Buonomo et al., 2000; Kitajima et al., 2003; Klein et al., 1999; Watanabe and Nurse, 1999).
Like yeast, the vertebrates analyzed to date have two Rad21/Rec8 proteins, with a similar partition of function (Lee and Orr-Weaver, 2001; Nasmyth, 2001). By contrast, higher plants such as Arabidopsis and rice (Bai et al., 1999; da Costa-Nunes et al., 2006; Dong et al., 2001; Liu and Makaroff, 2006; Zhang et al., 2006) have four Rad21/Rec8 proteins, only one of which (Syn1/DIF1 in Arabidopsis, OsRad21-4 in rice) (Bai et al., 1999; Bhatt et al., 1999; Cai et al., 2003; Zhang et al., 2006) has been identified as essential for meiosis. Identifying the biological function of the other three proteins is important for further understanding why higher plants contain four Rad21/Rec8 proteins.
Here we have characterized the function of OsRad21-3 by phylogenetic, gene expression, protein immunolocalization and RNAi-mediated reverse genetic analyses. Our results showed that OsRad21-3 is an orthologue of yeast Rad21. The OsRAD21-3 transcript is preferentially present in flowers, and displays timing of expression that is different from that of the other three OsRAD21 genes (Zhang et al., 2004, 2006, data not shown). Its expression in flowers persists until the mature pollen stage. Use of RNA interference to knock-down OsRAD21-3 expression led to arrested mitosis and aberrant chromosome behaviors in developing pollen. These results indicate that OsRad21-3 protein plays a role in pollen development.
Results
Characteristics of OsRad21-3
Our previous analyses predicted that the rice genome encodes four Rad21/Rec8-like proteins – OsRad21-1, OsRad21-2, OsRad21-3 and OsRad21-4 (Zhang et al., 2006). The full-length OsRAD21-3 cDNA (AK101268) of 2856 bp is capable of encoding a polypeptide of 728 amino acids with a calculated molecular mass of 80.2 kDa. A BLASTN search of the TIGR rice genomic database gave a hit for the full-length cDNA in chromosome 8 only (Figure S1a), which indicates that the gene is a single-copy gene. OsRAD21-3 cDNA showed no obvious similarity with any of the other three OsRAD21 cDNAs (Zhang et al., 2006).
Like OsRad21-1 and OsRad21-4 (Zhang et al., 2004, 2006) and other known Rad21/Rec8 proteins from other species (Zhang et al., 2004), OsRad21-3 consists of the complete Pfam04825 and Pfam04824 domains in its N-terminus (amino acids 1–110) and C-terminus (amino acids 669–723), respectively, and a long linker region between them. The middle linker sequence shows high variance in sequence and length compared with other Rad21/Rec8 proteins, and contains a potential bipartite nuclear localization signal (amino acids 378–394), a separase recognition site (amino acids 154–164) and two PEST motifs (amino acids 229–263 and 490–503). These motifs are functionally conserved in the known Rad21/Rec8 proteins (Nasmyth, 2001; Zhang et al., 2004). Consistent with these features, OsRad21-3 showed nucleus localization in a transient experiment with onion epidermic cells (Figure S1b).
OsRad21-3 displays only low similarity with the other three OsRad21s in rice (approximately 22% overall amino acid identity), and the similarity is limited mainly to the domain-containing N- and C-terminal regions. A comparison of sequence similarity between OsRad21-3 and the other three OsRad21s or Rad21/Rec8 proteins from other species revealed that OsRad21-3 was more similar to Arabidopsis Syn3 (also called AtRad21.2) than to the others. In addition, phylogenetic analysis based on Clustal W alignments of N-terminal Pfam04825 domain regions, which are highly conserved in all known Rad21/Rec8 proteins (Zhang et al., 2004), revealed that OsRad21-3 tended to form a group with known members of the Rad21 subfamily from yeast (Michaelis et al., 1997), Drosophila (Warren et al., 2000), Caenorhabditis elegans (Pasierbek et al., 2001), mice and humans (McKay et al., 1996). Together, these results suggest that OsRad21-3 is a member of the Rad21 subfamily.
OsRAD21-3 is expressed preferentially in flowers
Semi-quantitative RT-PCR revealed that OsRAD21-3 expressed preferentially in flowers, and weakly in leaves, buds and roots (Figure 1a). Furthermore, immunoblot analysis with antibody to OsRad21-3 revealed that the protein was most abundant in flowers and barely detectable in leaves, buds and roots (Figure 1b).
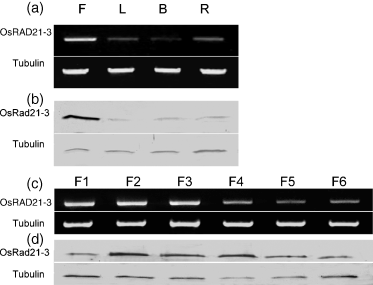
Expression profiles of the OsRAD21-3 gene.Expression of OsRAD21-3 in various tissues (a) and developing flowers (c) was analyzed by semi-quantitative RT-PCR. The PCR products were separated on 1% agarose gels. The amplified rice tubulin A mRNA was used as a constitutive control. For Western blot analysis (b, d), total proteins prepared from these materials were subjected to SDS–PAGE, and then immunoblotted using an antibody against OsRad21-3 or against tubulin (control).(a, b) Patterns of OsRAD21-3 mRNA (a) and protein (b) accumulation in flowers (F), leaves (L), buds (B) and roots (R).(c, d) Accumulation profiles of OsRAD21-3 mRNA (c) and protein (d) in developing flowers at the following stages: stamen and carpel primordial formation (F1), pollen mother cell formation (F2), male meiosis (F3), uninucleate microspore (F4), bicellular pollen (F5) or tricellular pollen (F6).
To further investigate the expression of OsRAD21-3 in developing flowers, we collected flowers at various developmental stages based on the length of flowers and young panicles, and performed cytological observations. Flowers that were ≤0.9 mm from panicles of <15 mm were grouped as F1 (stamen and carpel primordia formation stage), flowers of 0.9–2.5 mm in panicles of 15–50 mm as F2 (pollen mother cell formation stage), flowers of 2.5–5 mm in panicles of 50–100 mm as F3 (male meiosis stage), and flowers of 5–7 mm in panicles of >100 mm as F4 (microspore stage). Flowers that were ≥7 mm, with anthers becoming yellow in the panicles remaining before the heading stage, were grouped as F5 (bicellular pollen stage), while those close to anthesis were grouped as F6 (tricellular pollen stage). Unlike the OsRAD21-4 transcript, which is at its highest level at the meiosis stage and decreased in level as meiosis proceeds (Zhang et al., 2006), the level of OsRAD21-3 transcript peaked at F1–F3 and decreased thereafter (Figure 1c). Moreover, immunoblot analysis revealed that the level of OsRad21-3 protein peaked at F2–F4, and then decreased slightly (Figure 1d). The accumulation dynamics of OsRAD21-3 protein in these developing flowers are consistent with pollen development from meiocytes to tricellular pollen.
OsRAD21-3 is expressed in microspores and pollen grains
To determine whether OsRAD21-3 mRNA accumulates in male cells, we performed RNA in situ hybridization using DIG-labeled OsRAD21-3 antisense and sense RNA probes. In anthers, signals were detected in pre-meiotic pollen mother cells (PMCs) (Figure S2a), meiotic PMCs (Figure S2b) and microspores (Figure S2c), and were not detected in bicellular and tricellular pollen (data not shown). The signal was present in tapetal cells (Figure S2a–c). Failure to detect a signal in bicellular and tricellular pollen might result from a relatively low level of the transcripts in such pollen. Sense (control) probes did not produce signals in flowers (Figure S2d–f).
To clarify whether the transcript and protein are present in pollen grains, we performed immunoblot experiments using mature and in vitro-germinated pollen grains, and revealed that the protein was present in these pollen grains (Figure 2a). Combined with the data above, this result demonstrates that OsRAD21-3 is expressed in the male cells from PMCs to mature pollen stages.
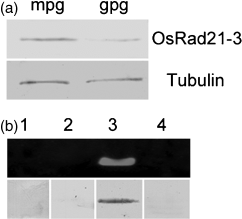
Examination of OsRAD21-3 transcript and protein in pollen grains.(a) Western blot detection of OsRad21-3 protein in mature (mpg) and germinated (gpg) pollen grains, with tubulin protein as a constitutive control, using a procedure identical to that described in Figure 1.(b) RT-PCR analyses of OsRAD21-1 (1), OsRAD21-2 (2), OsRAD21-3 (3) and OsRAD21-4 (4) transcripts (upper panel), and Western blot results for each corresponding protein (lower panel) in pollen grains.
As the rice genome has four OsRAD21 genes, we used RT-PCR and immunoblot analysis to examine whether transcripts and proteins of the other three genes are also present in pollen grains. As seen in Figure 2b, pollen grains contained transcript and protein of OsRAD21-3 only, not the other genes. Taken together, these results strongly suggest the involvement of OsRAD21-3 in pollen development.
OsRad21-3 localizes in mitotic chromosomes
Considering that the functionally characterized Rad21/Rec8 proteins of various species play roles in chromosome cohesion (Lee and Orr-Weaver, 2001; Nasmyth, 2001; Uhlmann, 2003), we used immunostaining to examine the chromosome localization of OsRad21-3 using an antibody against this protein in root tip cells. The antibody was developed using a non-conserved fragment of OsRad21-3 protein, which has no similarity with any of the other three OsRad21 proteins and did not cross-react with them (Figure S3a). Furthermore, the antibody detected only one band on Western blotting with total proteins prepared from flowers and vegetative tissues (Figure 1b,d, Figure S3b), and the band had an estimated molecular mass similar to the theoretical value for OsRad21-3 (Figure S3b). This result indicates that the antibody specifically recognizes OsRad21-3. Immunostaining experiments revealed that OsRad21-3 was localized on mitotic chromosomes (Figure 3a–c), but not in other parts of mitotic cells or meiotic cells (data not shown). The labeling signal was detected at interphase (Figure 3a), became strongest at pre-prophase (Figure 3b), decreased sharply at prophase (Figure 3c), and was not detectable at and after metaphase (Figure 3d). This result suggests that a large amount of OsRad21-3 is released during prophase. The selective release feature is similar to that for mitotic cohesin from vertebrates (Waizenegger et al., 2000), which suggests involvement of the protein in chromosome cohesion.
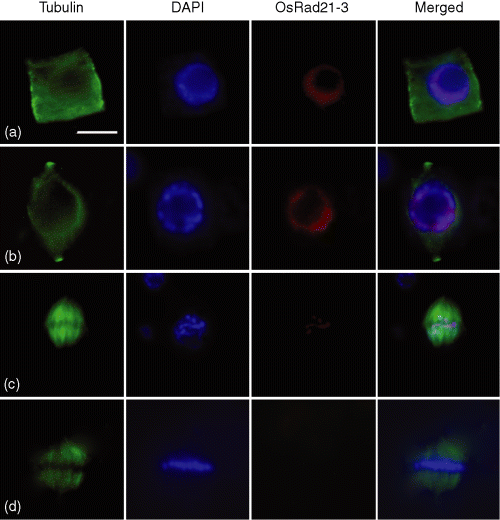
Immunofluorescent staining of OsRad21-3 in mitotic chromosomes from root tip cells of wild-type plants.Nuclear spreads were stained with anti-β-tubulin antibody (green) or with OsRad21-3 antibody (red), and counterstained with DAPI (blue). (a) Interphase; (b) pre-prophase; (c) prophase; (d) metaphase. Bar in (a) 5 μm, and applies to all images. The results are representative of at least 50 cells for each phase.
OsRAD21-3i lines show a decreased seed setting rate
We addressed the function of OsRAD21-3 using the RNA interference approach (Wesley et al., 2001). To guarantee the specificity of RNAi, we constructed the OsRAD21-3 RNAi vector pOsRAD21-3i using a 729 bp fragment of OsRAD21-3 cDNA (nt 1278–2006) that shares no similarity to the other three OsRAD21 cDNAs. We obtained 30 independent transformants from rice calli transformed with pOsRAD21-3i. A PCR check for transgene insertion using a primer pair localized in the spacer and inserted cDNA sequences in pOsRAD21-3i revealed the expected fragment of approximately 1 kbp in all 30 T0 transgenic plants but not in wild-type plants. Compared with the wild-type, OsRAD21-3i lines exhibited no detectable abnormalities during vegetative growth. Flowers from these RNAi lines were indistinguishable from those of wild-type. However, in contrast to the wild-type, which had a 90% seed-setting rate, 23 of the 30 lines had a greatly decreased seed-setting rate. Furthermore, we investigated the seed-setting rate of nine of the 23 lines using R1 plants regenerated from stalks of their T0 lines. All nine had a low seed-setting rate (10% in one line, 11–46% in six lines and 50–67% in two lines). This finding suggests that OsRAD21-3 is involved in fertility.
Finally, we examined the insertion copy number of the transgene in the nine lines using Southern blot hybridization, with wild-type plants as a control, and found two insertions in one line, one insertion in the remaining eight lines, and none in wild-type plants. Five RNAi lines (L15, L16, L17, L19 and L23) with a single-copy insertion and representing different sterile phenotypes (Figure 4a) were used for further analysis.
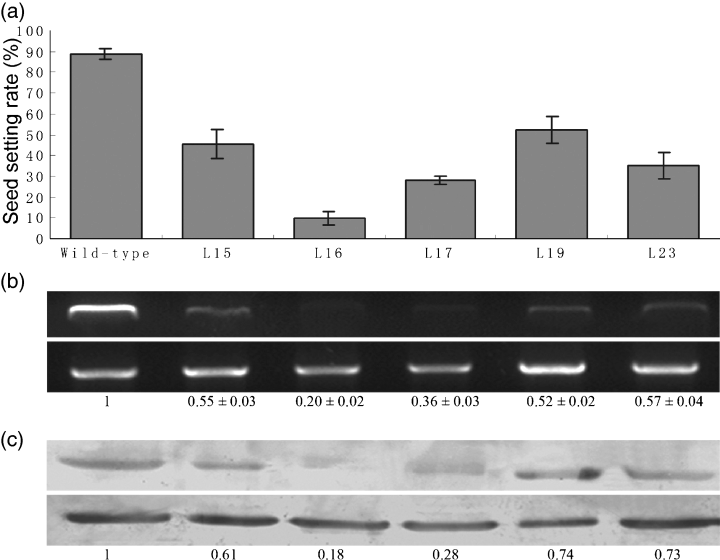
RNAi-mediated down-regulation of OsRAD21-3 transcript and protein correlates with sterility in OsRAD21-3i lines.(a) RNAi lines L15, L16, L17, L19 and L23 had severely decreased seed-setting rates compared with wild-type (t-test, P-value ≤ 0.01).(b) Endogenous OsRAD21-3 transcript was greatly down-regulated in the RNAi lines. Semi-quantitative RT-PCR was performed using total RNA extracted from panicles of the RNAi lines and wild-type and the primer pair P1 F/P1R. Top panel: OsRAD21-3. Bottom panel: constitutively expressed tubA. The quantified levels of OsRAD21-3 mRNA (data for RNAi lines normalized to that for wild-type) are shown below the panels.(c) OsRad21-3 protein was deficient in the RNAi lines. The endogenous protein level was detected by Western blotting of total proteins extracted from the panicles of the five RNAi lines and wild-type. Estimated OsRad21-3 protein levels (data for RNAi lines normalized to that for wild-type) are shown below the panels. Top panel: OsRad21-3. Bottom panel: tubulin control.
Downregulated OsRad21-3 protein in OsRAD21-3i lines correlates with their sterility
The double-stranded RNA with hairpin structures formed from the transcribed sense–spacer–antisense sequence in RNAi constructs gives rise to small RNAs and ultimately reduces or eliminates the endogenous transcript and protein of a target gene (Hamilton and Baulcombe, 1999). To investigate whether endogenous OsRAD21-3 transcript and protein were knocked down in the RNAi lines, we examined the levels of endogenous transcript and protein by RT-PCR and Western blot, respectively, in panicles of the L15, L16, L17, L19 and L23 lines, as well as wild-type plants. The endogenous transcript and protein were considerably down-regulated in the five RNAi lines compared with that in the wild-type (Figure 4b,c). We also examined small RNAs specific to OsRAD21-3 by Northern hybridization in the five RNAi lines, and found small RNAs in all five lines but not in the wild-type (data not shown), with the highest level in L16, which is consistent with the most severe down-regulation of the endogenous transcript and protein in the same line. Clearly, down-regulation of both transcript and protein in the RNAi lines is a factor in their sterile phenotypes (Figure 4a–c).
OsRAD21-3i lines show defects in pollen viability
Because pollen is a key regulator of successful fertilization in sexual plants, and because OsRAD21-3 transcript and protein are present in microspores and pollen grains, we first examined the viability of mature pollen grains of the five RNAi lines and wild-type plants using I2-KI and Alexander staining (Alexander, 1969). Starch is preferentially stored in rice pollen and metabolized upon germination to supply a carbon skeleton and energy for the growing pollen tube, and is thus an important indicator of the viability of the pollen (Dai et al., 2006). I2-KI staining (Figure S4a,b) revealed that more than 95% of wild-type pollen grains (n = 1263) were round and had a uniform size, and showed with a dark blue–black reaction, whereas pollen grains from the RNAi lines were variable in size, being smaller or larger than the wild-type, with a greatly decreased number being stained, ranging from 28% (n = 1671, in L16) to 75% (n = 1580, in L19). Alexander staining (Figure S4c,d) revealed that 95% of wild-type pollen grains (n = 1230) were ‘viable’ (red), whereas approximately 40% (n = 1660) of those from L16 appeared ‘viable’ and the remainder were ‘non-viable’ (not stained); similar results were observed for the other four lines, in accordance with the I2-KI assay results.
The staining assays gave only an indirect estimation of pollen viability, so to examine pollen viability directly, we performed in vitro pollen germination experiments. Under the assay conditions (Figure S4e,f), 83% of wild-type pollen grains (n = 644) germinated, but germination was observed in only 10% of L16 pollen grains (n = 821), approximately 30% of L17 pollen grains (n = 756), 23% of L23 pollen grains (n = 766), and approximately 40% of L15 (n = 657) and L19 (n = 712) pollen grains (Figure 5). Together, these results indicate that pollen viability is disrupted in the RNAi lines.
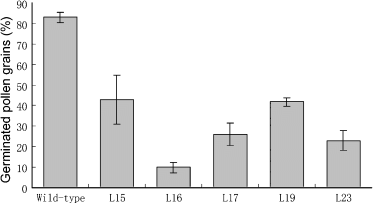
Pollen viability is disrupted in OsRAD21-3i lines.The viability of pollen grains (percentage) at the mature pollen stage from wild-type and RNAi plants was examined by in vitro germination. The data are the means ± SD of three biological repeats.
Further observations revealed that, although pollen grains from the RNAi lines were variable in size, as compared with the uniform size of wild-type pollen grains, most were round or pear-shaped and only a few were shrunken. Using the method described by Zhang et al. (2006), we estimated the pollen amount per anther in these mutants and found no apparent difference compared to the wild-type, which suggests that microsporogenesis might be not disrupted severely in the RNAi lines, and that sterility is mainly derived from abnormal pollen development at the post-meiotic stage.
Male meiosis was not severely disrupted in OsRAD21-3i plants
As the OsRAD21-3 transcript was detectable in male meiocytes, and to clarify the preceding findings, we observed meiosis behaviors using male meiocytes from the L16, L17 and L23 lines as well as wild-type plants. In wild-type male meiocytes, after synapsis and condensation, homologous chromosomes formed 12 bivalents at diakinesis (Figure 6e) and aligned at the equatorial plate at metaphase I (Figure 6f). The bivalents then underwent reductional division at anaphase I (Figure 6g), generating dyads (Figure 6h). During meiosis II, the two daughter cells divided simultaneously, with two parallel orientations of spindles (Figure 6i), generating tetrads.
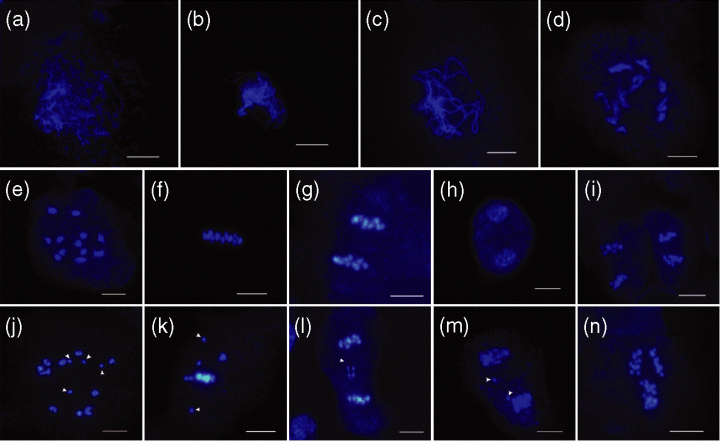
Male meiosis in wild-type and OsRAD21-3i plants.Nuclear spreads were prepared from fixed anthers from wild-type plants (e–i) and OsRAD21-3i lines (a–d, j–n) and stained with DAPI. Scale bar = 10 μm.(a–d) Prophase I at leptonema (a), zygonema (b), pachynema (c) and diplonema (d) in the RNAi plants appeared normal.(e–i) Wild-type male meiosis. (e) Diakinesis, (f) metaphase I, (g) anaphase I, (h) dyads, (i) metaphase II.(j–n) Male meiocytes from the RNAi lines. (j) Diakinesis, showing univalents (arrowheads); (k) metaphase I and (l) anaphase I, showing lagging chromosomes (arrowheads); (m) early dyads, showing small nuclei (arrowheads); (n) anaphase II, showing the non-parallel plates in two daughter cells.
In male meiocytes from the RNAi lines (data shown only for L16), meiosis appeared to follow that of the wild-type from leptotene to diplotene (Figure 6a–d). Condensation and synapsis of chromosomes occurred with no noticeable differences compared to the wild-type. At diakinesis, most male meiocytes had 12 recognizable bivalents; only 9% of cells (n = 80) had univalents (Figure 6j). The desynapsed homologous chromosomes did not align at the metaphase plate (approximately 7%,n = 55) (Figure 6k). As a result, lagging chromosomes appeared at anaphase I (Figure 6l,m) in some male meiocytes (approximately 5%, n = 60). During meiosis II, abnormally oriented plates in two daughter cells occurred in <3% of male meiocytes (Figure 6n). Lines L17 and L23 showed less than 5% of male meiocytes with aberrant chromosome behaviors. At the tetrad stage, only a few spores from these RNAi lines appeared to have small nuclei or aberrant size, and the appearance of most (90–95%, n = 50) was similar to that of the wild-type. These observations demonstrate that male meiosis was not severely disrupted in the RNAi lines.
OsRAD21-3i lines showed aberrant pollen development at the post-meiotic stage
On the basis of the above results, we examined immature and mature pollen grains from the five RNAi lines as well as from wild-type plants by DAPI staining. After being released from tetrads, wild-type microspores increase in size (Figure 7a,b), with the nucleus migrating to the side opposite to the aperture (Feng et al., 2001; Owen and Makaroff, 1995). This process ends with initiation of asymmetric PMI (Figure 7c), whereby microspores develop into bicellular pollen grains with a dispersed nucleus in the large vegetative cell and a condensed nucleus in the small generative cell (Figure 7d), which then generates two sperm cells by PMII. Thus, mature pollen grains in rice are tricellular (Figure 7e).
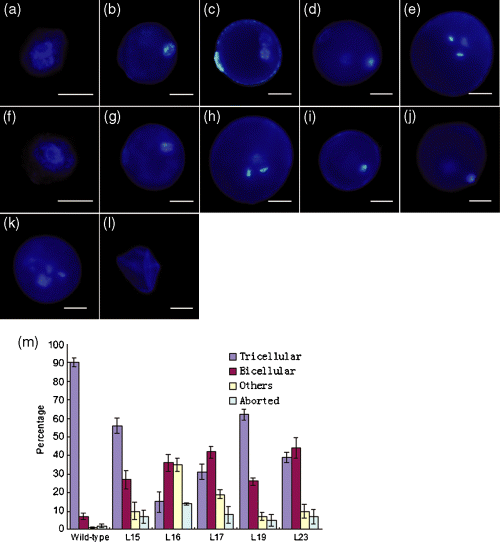
Abnormal pollen development in OsRAD21-3i plants.Pollen development from uniuncleate microspores to tricellular pollen in wild-type plants (a–e) and RNAi lines (f–l) by DAPI staining. Scale bar = 12.5 μm.(a–e) Wild-type pollen. (a) Early uninucleate microspore, (b) late uninucleate microspore, (c) early bicellular pollen, (d) mid to late bicellular pollen, (e) mature pollen.(f, g) Uninucleate microspores from the RNAi lines. (f) Early uninucleate microspore and (g) late uninucleate microspore, showing normal appearance.(h–l) Pollen grains in the RNAi lines at the mature pollen stage corresponding to the mature pollen phase of wild-type plants. (h) Mature pollen with wild-type tricellular phenotype, (i) arrested unicellular pollen, (j) arrested bicellular pollen, (k) pollen with more than three nuclei, (l) aborted pollen.(m) Frequencies of normal and aberrant pollen grains at the mature pollen stage in wild-type and RNAi plants. ‘Others’ include arrested uninucleate microspores and pollen with more than three DAPI-stained loci. The data are the means ± SD of three biological repeats. Compared with the wild-type, RNAi lines showed aberrant pollen development (t-test, P-values < 0.05).
Most microspores from OsRAD21-3i lines (90–95%, n = 200) (Figure 7f,g) were indistinguishable from those of the wild-type (Figure 7a,b). However, at the mature pollen stage, at least 90% of pollen grains from the wild-type were tricellular (n = 1663) (Figure 7e,m), with about 7% bicellular and 3% aborted (undetectable nucleus and shrunken) (Figure 7m); in contrast, only 15% of mature pollen grains from L16 were tricellular (n = 2130) (Figure 7h,m), 71% had an aberrant nucleus appearance (Figure 7i–k,m) and 14% aborted (Figure 7l,m). These defective pollen grains with aberrant nucleus appearance were arrested in the uninucleate (Figure 7i) or binucleate (Figure 7j) stage, or had more than three DAPI-stained loci (Figure 7k) and were smaller than wild-type tricellular pollen (Figure 7i–k). A similar situation was found in the other four lines (Figure 7m). The nucleus in the arrested unicellular pollen (Figure 7i) was abnormally condensed. The arrested bicellular pollen grains had a wild-type bicellular pollen-like appearance (Figure 7j versus Figure 7d) that is indicative of the completion of PMI but failure of PMII. All these observations indicate that the OsRad21-3 deficiency in the RNAi lines leads to arrest in mitosis and aberrant chromosome behaviors during pollen development.
The quantified endogenous protein level and the proportion of normal tricellular pollen grains in the RNAi lines and wild-type plants were strongly correlated (r = 0.911). Together, our results suggest that the OsRAD21-3 deficiency in the mutants resulted in disruption of post-meiotic pollen development.
Discussion
OsRad21-3 is an orthologue of yeast Rad21
Yeast has two members in the Rad21/Rec8 family, Rad21 and its meiotic variant Rec8. Rad21 functions mainly in mitotic cohesion and Rec8 specifically in meiotic cohesion (Klein et al., 1999; Michaelis et al., 1997; Watanabe and Nurse, 1999). This situation is also found in the vertebrates analyzed to date (Lee and Orr-Weaver, 2001; Nasmyth, 2001). By contrast, rice and Arabidopsis have four members of the family, all sharing motif/domain features with the known Rad21/Rec8 proteins from other species (Bai et al., 1999; da Costa-Nunes et al., 2006; Zhang et al., 2004, 2006). However, only one (OsRad21-4 in rice, Zhang et al., 2006; Syn1 in Arabidopsis, Bai et al., 1999) has been identified as essential for meiosis.
In Arabidopsis, the three other RAD21-like genes show overlapping profiles of expression in a wide variety of tissues (da Costa-Nunes et al., 2006). The function of SYN3/AtRAD21.2 remains unknown, but single or double mutants of the other two genes showed normal growth and development, which suggests a redundant function of the three genes, at least in normal growth (da Costa-Nunes et al., 2006). Our results show that OsRad21-3 is an orthologue of yeast Rad21. OsRAD21-3 is expressed in vegetative tissues, albeit at a much lower level than detected in flowers. However, OsRAD21-3i lines did not display obvious defects in vegetative growth. A previous study identified OsRAD21-1 as another rice orthologue of yeast RAD21 (Zhang et al., 2004). OsRAD21-3 shows an expression profile that overlaps with that of OsRAD21-1 in vegetative tissues (Zhang et al., 2004; this paper). Based on these data, OsRAD21-3 possibly has functional redundancy with OsRAD21-1 in sporophytic cells.
Possible roles of OsRAD21-3 in meiosis
OsRAD21-3i lines displayed aberrant meiotic phenotypes in a low proportion of male meiocytes from late prophase I to meiosis II. Consistent with this finding, the average number of pollen grains per anther in these lines was not decreased significantly compared with the wild-type. By contrast, the Arabidopsis syn1 mutant and OsRAD21-4 RNAi lines show severe meiotic defects from early prophase I (Bai et al., 1999; Zhang et al., 2006). The defects in early prophase I meiosis in these two mutants and in other plant meiotic mutants (Li et al., 2005; Nonomura et al., 2004; Pawlowski et al., 2004; Stevens et al., 2004) usually leads to a sharp decrease in male spore number because aberrant spores derived from the defects die early. Based on these observations, OsRAD21-3 is possibly implicated in meiosis but functions mainly from late prophase I to meiosis II. Alternatively, because OsRAD21-3i lines contained residual OsRad21-3 protein, we cannot rule out the possibility that the residual OsRad21-3 in male meiocytes is sufficient for early meiosis. In fission yeast and mouse, RAD21 genes have been identified as involved in meiosis, in addition to having important roles in mitosis (Watanabe and Nurse, 1999; Prieto et al., 2002; Prieto et al. 2004; Xu et al., 2004).
However, inconsistent with these notions is the fact that the immunostaining experiments described here did not show signals for OsRad21-3 protein in male meiotic chromosomes. Several studies have suggested that chromosome-bound cohesin protein is undetectable at low levels (Cai et al., 2003; Liu and Makaroff, 2006) and/or that the specific chromatin configuration in some domains blocks access of antibodies against cohesin proteins (Liu and Makaroff, 2006; Tomonaga et al., 2000) for immunostaining. In mammalian cells, well-known mitotic cohesin proteins such as STAG2 and Rad21 were detectable in meiotic chromosomes using antibodies against short peptides but not by antibodies against polypeptides of these proteins (Prieto et al., 2002), which implies that these factors may affect immunostaining detection of OsRad21-3 in meiotic chromosomes. Further experiments using an antibody against short peptides from OsRad21-3 are required to clarify this.
OsRAD21-3 is required for post-meiotic development of pollen
In contrast to male meiosis, pollen development at the post-meiotic stage was severely disrupted in OsRAD21-3i lines, which showed arrested uninucleate and binucleate pollen and abnormal chromosome segregation. Together with the expression of OsRAD21-3 in developing pollen, this result clearly indicates that OsRAD21-3 is necessary for pollen development. These phenotypes were reminiscent of those found in Rad21-deleted vertebrate cells (Sonoda et al., 2001) and the yeast rad21 mutant (Guacci et al., 1997). In these mutants, aberrant cohesion between sister chromatids contributes to defects in nuclear division and chromosome behaviors (Guacci et al., 1997; Sonoda et al., 2001). Although it difficult to follow the various phases of PMI and PMII (Chen and McCormick, 1996; Durbarry et al., 2005; Howden et al., 1998; Iwakawa et al., 2006), given that the mitotic chromosome localization for OsRad21-3 is similar to that for the Rad21 proteins from yeast and vertebrates (Guacci et al., 1997; Sonoda et al., 2001), we can propose that the role of OsRad21-3 in pollen development results mainly from its involvement in sister chromatid cohesion.
Rad21-containing cohesin coordinates sister chromatid behaviors in cell division by holding them together (Lee and Orr-Weaver, 2001; Nasmyth, 2001); and the cohesin in vertebrates and yeast has been found to have roles in congregating paired sisters into a metaphase plate and in subsequent bipolar attachment of sister kinetochores to a spindle (Uhlmann, 2003). Rad21-deleted yeast and vertebrate cells fail to align chromosomes at the metaphase plate and to establish stable attachment of sister kinetochores to a spindle. The failure finally leads to arrested nuclear division by a spindle checkpoint pathway, which functions to guarantee faithful segregation of chromosomes (Sonoda et al., 2001; Toyoda et al., 2002; Uhlmann, 2003). This possibly explains why OsRad21-3 deficiency in RNAi lines causes arrest in pollen mitosis.
Because microspore isolation after meiosis leads to the independent development of individual spores (Twell, 2002), there are two possible explanations for the phenotypic variation observed in the defective spores of OsRAD21-3i plants. The first is that different amounts of residual OsRad21-3 protein are distributed into the spores after meiosis. The presence of a small number of tricellular pollen grains in these RNAi lines indirectly supports this explanation. Alternatively, the de novo synthesis of OsRad21-3 in the spores may be disrupted at different levels.
In conclusion, this study shows that, of the four OsRAD21 genes in the rice genome, only OsRAD21-3 is expressed in pollen grains and is required for pollen development. Our findings show, in part, why the much-analyzed flowering plants rice and Arabidopsis have four Rad21/Rec8 proteins compared with only two in yeast and metozoans, and give some clues to understanding the functional differentiation of Rad21/Rec8 proteins in evolution.
Experimental procedures
Plant materials and extraction of nucleic acids
Seedlings of cultivar Zhonghua 10 (Oryza sativa L. ssp. japonica) were planted in soil and raised to maturity. Total RNA extracted various tissues using a Trizol kit (Invitrogen; http://www.invitrogen.com/) was treated with RNase-free DNase I (TaKaRa, http://www.takara.com.cn) to remove residual genomic DNA.
OsRAD21-3 cDNA cloning
A partial cDNA sequence of OsRAD21-3 was obtained by RT-PCR using gene-specific primers P1 F (5′-ATGTTCTACTCGCACACG-3′) and P1R (5′-CCATAGGCTGCTTCTTGT-3′). This amplified fragment is 2139 bp (accession number AY371048). A full cDNA sequence (accession number AK101268) that has recently been deposited into GenBank is identical to the above sequence, with an additional extension at the 5′ and 3′ ends, and represents the full-length cDNA sequence of OsRAD21-3; this sequence was thereafter used in this study.
Expression analysis
Accumulation patterns of OsRAD21-3 mRNA in various tissues were analyzed by semi-quantitative RT-PCR with primers P1 F and P1R. The transcript of the tubulin tubA gene (accession number X91806) was used as a constitutive control (Ding et al., 2002). PCR was performed for 25 cycles. To determine the expression profiles of the gene in various flower cells by in situ hybridization, flowers were fixed and further processed as described previously (Ding et al., 2002; Meyerowitz, 1987). DIG-labeled sense and antisense RNAs used for hybridization were synthesized by in vitro transcription (Roche, http://www.roche-applied-sciences.com) using a 184 bp cDNA fragment spanning sites 940–1148 of the OsRAD21-3 cDNA, which shows no similarity to the other three rice OsRAD21 genes, and was amplified using primers P2 F (5′-CCCTGAACTTAACTTGCC-3′) and P2R (5′-AGATAATCCACCTTGACC-3′).
Polyclonal antibody production and Western blotting
A 729 bp fragment of OsRAD21-3 cDNA corresponding to amino acids 382–623, which shares no similarity to the other three OsRad21 genes, was amplified with primers P3 F (5′-TAGGATCCATTGATGGCGGTGAACTAC-3′, addedBamHI site underlined) and P3R (5′-TAGTCGACGAATCTCGGGCAAATCTC-3′, added SalI site underlined), cloned into pET-28a(+) (Novagen, http://www.emdbiosciences.com), and confirmed by sequencing. Escherichia coli DH5α cells carrying the resulting plasmid were cultured to generate the polypeptide fragment of the OsRad21-3 protein. The recombinant protein purified by Ni2+ affinity chromatography (Qiagen; http://www.qiagen.com/) was used for generation of a polyclonal rabbit antibody against OsRad21-3.
Total protein preparation from various tissues and Western blot detection of OsRad21-3 were conducted according to the methods described by Zhang et al. (2006). The primary antibody used was that against OsRad21-3 (1:4000 dilution) or tubulin (1:5000 dilution, Sigma; http://www.sigmaaldrich.com/). The antigen–protein complex was detected using goat anti-rabbit IgG-conjugated alkaline phosphatase.
Subcellular localization and fluorescence immunostaining
The whole ORF of OsRAD21-3 amplified using primers P4 F (5′-TATGGATCCGATGTTCTACTCGCACAC-3′) and P4R (5′-AGTACTAGTTTTGGCTCCTGA GAGTGA-3′), with engineered BamHI and SpeI enzyme sites at the 5′ and 3′ ends, respectively, was inserted into pCAMBIA1302 (CAMBIA, http://www.cambia.org), and confirmed by sequencing. The construct 35S::GFP:OsRAD21-3 was bombarded into onion epidermis using the biolistic PDS-1000/He particle delivery system (Bio-Rad; http://www.bio-rad.com/). GFP signals were observed under a microscope with a FITC filter (Zeiss; http://www.zeiss.com/).
Immunolocalization of OsRad21-3 was performed according to the method described by Liu et al. (1993). Nuclear spreads were first incubated with primary anti-OsRad21-3 antibody (1:5000 dilution) in a blocking buffer pH 7.4 (5% BSA, 5% goat serum and 0.1% Tween-20 in 1× PBS buffer) at 4°C overnight, and then with TRITC-labeled goat anti-rabbit IgG (Jackson ImmunoResearch Laboratories, http://www.jacksonimmuno.com) (1:100 dilution) or with fluorescein-labeled mouse anti-β-tubulin monoclonal antibody only (Sigma) at room temperature for 2–3 h. Finally, the spreads were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) solution. Signals were detected using a Zeiss fluorescence microscope with two, four and five filters for DAPI, fluorescein and TRITC, respectively. Images obtained by cool CCD were merged using Image Pro-Plus 5.0 software (Media Cybernetics, http://www.mediacy.com).
RNAi vector construction and plant transformation
The OsRAD21-3 RNAi vector was constructed by inserting a 729 bp fragment of OsRAD21-3 cDNA into pWTC605, an RNAi tool vector derived from pCAMBIA1300 (CAMBIA) (Zhang et al., 2006). First, the cDNA fragment was amplified using primer pairs, either P3 F and P5R (5′-AAGGTACCGAATCTCGGGCAA ATCTC-3′, KpnI site underlined) or P5 F (5′-AAGAGCTCTTGATGGCGGTGAACT AC-3′, SacI site underlined) and P3R (sequences of P3 F and P3R are given above). Finally, the two digested fragments were ligated into pWTC605 in the sense and antisense orientations, respectively.
The resulting RNAi construct, pOsRAD21-3i, was used to transform rice embryonic calli induced from mature embryos of cultivar Zhonghua 10, by the Agrobacterium tumefaciens EHA105-mediated method, to generate OsRAD21-3 RNAi (OsRAD21-3i) plants, as described by Hiei et al. (1994). The regenerated plants from hygromycin-resistant calli were transplanted to soil and grown under standard conditions.
Nuclear and cell spreads
Male meiocyte spreading and DAPI staining were performed according to the method described by Zhang et al. (2006). DAPI-stained meiotic chromosomes were observed under a fluorescence microscope (Zeiss Axioskop40, http://www.zeiss.com). For examination of nuclei in microspores and bi/tricellular pollen grains, flowers at the microgametogenesis stage were fixed in ethanol/acetic acid (3:1) solution. Microspores or pollen grains released by squashing the fixed anthers on a slide were stained by DAPI (Park et al., 1998). Each test was repeated three times, and the total number of counted pollen grains was more than 2000 in each case.
Examination of pollen viability and in vitro germination
For examination of pollen viability, mature pollen grains were stained in Alexander solution (Alexander, 1969) or 0.1% I2-KI solution and viewed by light microscopy. The quantified value was the mean of results from three independent experiments.
For in vitro germination, mature pollen grains collected at anthesis were immediately transferred into pollen germination medium (0.004% H3BO3, 3 mm Ca(NO3)2·4 H2O, 10% PEG-4000, 20% sucrose, 3 mg l−1 VB1), cultured at room temperature for 10 min, and examined under a light microscope. A pollen grain with a growing pollen tube was considered germinated. The pollen germination rate for each line was calculated using three independent biological samples, and each observation involved examination of at least 600 pollen grains.
Acknowledgements
We thank Zhukuan Cheng at the Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, for technical assistance with fluorescence immunostaining, and Cheng Yun at the Institute of Botany, Chinese Academy of Sciences, for assistance with microscope observations. This research was supported by the National Science Foundation of China (grant number 30570147) and the Chinese Ministry of Science and Technology (grant number 2006CB910105 and 2005CB120802).




