Global transcriptional upregulation in the absence of increased translation in Chlamydia during IFNγ-mediated host cell tryptophan starvation
Summary
The developmentally regulated intracellular pathogen Chlamydia pneumoniae is a natural tryptophan auxotroph. These organisms survive tryptophan starvation induced by host cell activation with IFNγ by blocking maturation to the infectious form. In most bacteria, the stringent response is induced during amino acid starvation to promote survival. However, the response of obligate intracellular pathogens, which are predicted to lack stringent responses to amino acid starvation, is poorly characterized. Chlamydial transcription and translation were analysed during IFNγ-mediated tryptophan starvation using genomic normalization methods, and the data revealed the novel findings that: (i) global chlamydial transcription was upregulated; and (ii) protein synthesis was dramatically reduced. These results indicate a dysregulation of developmental gene expression and an uncoupling of transcription from translation. These observations represent an alternative survival strategy for host-adapted obligate intracellular bacterial pathogens that have lost the genes for stringent control during reductive evolution.
Introduction
Chlamydia are developmentally regulated obligate intracellular bacterial pathogens that alternate between two functionally and morphologically distinct developmental forms during a productive growth cycle: infectious, metabolically inert elementary bodies (EBs) and non-infectious, metabolically active reticulate bodies (RBs) [reviewed in the study by AbdelRahman and Belland (2005)]. The molecular mechanisms required for chlamydial differentiation are poorly defined because of the lack of a tractable genetic system, but complete genome sequences for several species coupled with transcriptional studies have aided the characterization of developmental events. Gene expression patterns in Chlamydia can be broadly classified into three groups: early cycle (important for EB-to-RB differentiation), mid-cycle (RB growth and division) and late cycle (critical for RB-to-EB differentiation and preparation for early events after EB attachment) (Shaw et al., 2000).
Chlamydia fail to complete their developmental cycle when starved for nutrients such as amino acids and iron (Coles et al., 1993; Raulston, 1997) or when exposed to antibiotics such as penicillin (Matsumoto and Manire, 1970). Under these conditions they enter a semidormant survival state termed persistence. Persistence is characterized by viable, non-cultivable growth, by blockage of EB maturation, and by the presence of morphologically aberrant RBs that fail to divide but remain metabolically active (Beatty et al., 1994). It is unclear how gene regulation is altered during persistence, but late-stage genes are predicted to be downregulated because of the block in the progression of RBs to EBs. Persistence is reversible when the eliciting conditions are removed, and aberrant RBs resume normal growth with the subsequent production of infectious EB progeny (Beatty et al., 1995). Persistence may have in vivo relevance as chronic sequelae from chlamydial diseases is linked to persistence through identification of chlamydial antigens and nucleic acids in the absence of cultivable organisms from the site of disease (Theijls et al., 1991; Campbell et al., 1993).
Host cell nutrient limitation is a trigger for the chlamydial persistent growth phenotype. Many bacteria engage the stringent response to alleviate amino acid starvation stress through downregulation of stable RNA synthesis and upregulation of amino acid biosynthetic and protein degradation pathways (Chatterji and Ojha, 2001). The response of obligate intracellular pathogens to amino acid starvation is poorly characterized, although it is known that they have lost functional homologues of the genes required for the stringent response during adaptation to intracellular growth (Mittenhuber, 2001).
IFNγ is a critical component of immunity to intracellular pathogens. This immune-regulated cytokine augments antigen processing and presentation and induces a variety of host cell antimicrobial responses including tryptophan starvation in human cells (Pfefferkorn, 1984; Byrne et al., 1986). Chlamydia pneumoniae (Cpn) and Chlamydia trachomatis (Ctr) alter gene and protein expression and enter the persistent growth state during host cell IFNγ activation (Beatty et al., 1993; Shaw et al., 1999; Mathews et al., 2001; Pantoja et al., 2001; Belland et al., 2003a; Goellner et al., 2006; Mukhopadhyay et al., 2006; Polkinghorne et al., 2006). Therefore, we investigated the global chlamydial stress response to tryptophan starvation mediated by IFNγ. We quantified chlamydial gene expression normalized to DNA content over a time-course of treatment and reactivation to characterize chlamydial intracellular development in IFNγ-activated host cells. We found that late-stage (i.e. RB-to-EB differentiation) gene transcription was upregulated between 24 h and 48 h post infection (p.i.) during IFNγ-mediated persistence. This was surprising as these transcripts encode proteins only found on EBs and EBs are not produced under these conditions. Treatment of host cells with penicillin also elicits morphologically aberrant RBs that are unable to complete a productive infection cycle, but penicillin-induced persistence did not show upregulation of late gene transcripts. A microarray analysis of all open reading frames (ORFs) and intergenic regions revealed that chlamydial transcription in IFNγ-treated cells was globally upregulated, representing an unusual response to stress by a bacterium. Unexpectedly, protein synthesis decreased over the time frame when transcription was found to be globally upregulated, indicating that transcription and translation are uncoupled during IFNγ-mediated persistence. This study comprehensively describes the amino acid starvation response of an obligate intracellular pathogen and may reflect an alternate strategy for bacteria during amino acid starvation or in the absence of a stringent response.
Results
IFNγ treatment results in non-cultivable growth that is reversible
To verify that blockage in RB-to-EB maturation occurred during IFNγ treatment, a one-step growth curve was generated by monitoring recoverable inclusion forming units (IFUs) over time. We observed a decrease in IFUs (a proxy for EBs) between 0 h and 24 h p.i., reflecting the conversion of infectious EBs to non-infectious RBs (Fig. 1A). Untreated samples displayed a characteristic logarithmic increase in IFUs after 24 h p.i., as expected during productive growth, whereas IFU production was inhibited by greater than 99.9% in IFNγ-treated host cells. The failure of chlamydiae to mature into EBs in the IFNγ-treated cultures was confirmed by transmission electron microscopy. In untreated cultures large RB-containing inclusions were detected at 48 h p.i. and by 96 h p.i. inclusions contained predominantly EBs (Fig. 1D). In IFNγ-treated cells only small inclusions with aberrant chlamydial forms were detected at all time points tested (Fig. 1D) (Pantoja et al., 2001). When IFNγ-containing medium was removed and replaced by fresh medium with excess tryptophan at 48 h p.i. (i.e. reactivated), IFUs were recovered by 96 h p.i., demonstrating that the block in EB maturation by IFNγ was reversible, confirming work performed by others (Fig. 1A) (Beatty et al., 1995). We also examined the effects of adding penicillin to the culture medium after removal of IFNγ and addition of excess tryptophan. Under this condition penicillin blocked IFU production by greater than 99% compared with the reactivated controls (Fig. 1A), with extremely large, aberrant RBs predominating (Fig. 1D). These results suggested a marked ultrastructural difference between IFNγ- and penicillin-induced persistence [see also the study by Matsumoto and Manire (1970)].
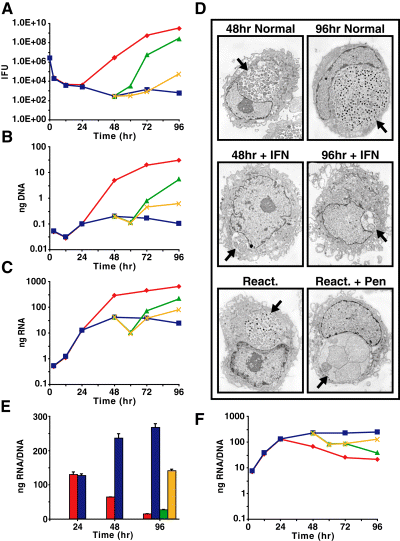
IFNγ-treatment of Cpn-infected cell cultures results in a persistent but reversible growth state. Cells were infected and treated with or without IFNγ at the time of infection. A subset of samples was reactivated at 48 h p.i. in the presence or absence of penicillin.A. Recoverable inclusion forming units presented as the average of three replicates for each sample.B and C. B. Quantification of chlamydial genomic DNA over a time-course. C. Quantification of chlamydial 16S rRNA over a time-course. A minimum of three replicate experiments were performed with standard deviations less than 5% of the sample.D. Electron micrographs of Cpn infected cells under conditions listed. Arrows indicate chlamydial inclusions.E and F. 16S rRNA transcripts plotted per genomic equivalent.Normal ( ); IFNγ (▪); reactivated (
); IFNγ (▪); reactivated ( ); reactivated with penicillin (
); reactivated with penicillin ( ).
).
We quantified chlamydial numbers under the different growth conditions by quantitative polymerase chain reaction (qPCR) of DNA because RBs and persistent chlamydial forms are not infectious, and therefore it is not possible to perform IFU determinations. We found that genomic DNA levels roughly paralleled the IFU numbers reported in Fig. 1A, including a failure to detect a significant increase in genomic DNA in the IFNγ-treated cultures after 24 h p.i. (Fig. 1B). Our results verified other published reports (Kane et al., 1999; Wood et al., 2003), suggesting tryptophan limitation mediated by IFNγ was not complete until after 24 h p.i., at which time DNA replication, chlamydial cell division and developmental cycle progression stopped. Our finding that DNA synthesis ceased was unexpected because a previous report showed that transcription of genes for DNA replication continues in the presence of IFNγ (Byrne et al., 2001).
Genomic DNA is the best parameter for normalization of transcription data
Stable RNA species (primarily 16S rRNA) are typically used to normalize transcriptional data in prokaryotic studies because stable RNA synthesis, as opposed to any given mRNA transcript or set of transcripts, is thought to be proportional to the chromosome (or growth rate) under all conditions. Early studies on the chlamydial developmental cycle found that the ratio of ribosomal RNA to DNA was higher in RBs than in EBs (Tamura, 1967). A more recent study concluded that 16S rRNA transcript levels are appropriate for normalization because 16S rRNA transcripts qualitatively mirrored genome accumulation (Mathews et al., 1999). However, no studies have been conducted to address whether 16S rRNA levels are appropriate for normalizing transcript data during persistence. Furthermore, bacteria respond to amino acid starvation by limiting stable RNA synthesis (Chatterji and Ojha, 2001). We therefore re-examined normalization to establish the most reliable method. Firstly, we found reduced accumulation of 16S rRNA after 24 h p.i. as RBs commenced conversion to EBs during the normal developmental cycle, as expected (Fig. 1C). Secondly, we found 16S rRNA transcription slowed even more dramatically in IFNγ-treated cultures (Fig. 1C), which was expected considering the reduced level of DNA replication and lack of cell division in these cultures (Fig. 1A,B and D). When we examined the ratio of 16S rRNA to DNA, we found the ratio dropped dramatically from 24 to 96 h p.i. during the normal developmental cycle, consistent with previous observations (Gutter and Becker, 1972). The ratio remained elevated in the IFNγ-treated infected culture (Fig. 1E and F), revealing a greater than 10-fold difference between normal and IFNγ-induced persistent growth by 96 h p.i. (Fig. 1E). Differences were also noted between the IFNγ-treated samples and samples reactivated in the presence or absence of penicillin (Fig. 1E and F). Based on these observations, the use of 16S rRNA to normalize transcription data under different growth conditions is less preferable to normalization based on DNA content (genomic equivalents) because: (i) the chromosome is the template for transcription and any variation in genome number should result in a proportional variation in transcript number; and (ii) DNA content best reflects the number of organisms under different growth conditions and during the developmental cycle. This method of normalization, while more involved than the use of rRNA because it requires isolation of DNA and RNA, may have general applicability for other prokaryotes.
Chlamydia pneumoniae transcriptional patterns during IFNγ-mediated persistence differ from developmentally regulated expression
Quantitative reverse transcription polymerase chain reaction (qRT-PCR) analyses were performed using a panel of Cpn genes that are associated with early, mid and late cycle events to monitor gene expression changes for these classes of genes during IFNγ-mediated tryptophan starvation (Fig. 2A–G). We chose euo, a gene encoding a DNA binding protein (Zhang et al., 1998), as our early gene; it was most highly expressed relative to other genes shortly after infection (12 h p.i.; Fig. 2A), and other stage-specific transcription studies have identified euo as a prototype early-stage gene (Shaw et al., 2000; Belland et al., 2003b). Our late gene panel consisted of omcB, encoding an EB-specific cysteine-rich outer envelope complex protein, hctA and hctB, encoding chromosome-compacting histone-like proteins, and lcrH_1, encoding a chaperone for type III secreted components. During a normal developmental cycle, transcription of late genes increased after 24 h p.i. as RBs began to condense into EBs (Fig. 2D–G). We chose two well-studied and highly transcribed genes to represent the mid-cycle: groEL_1, which encodes a 60 kDa heat-shock chaperone, and ompA, which encodes the major outer membrane protein. Transcription of mid-cycle genes is highest during RB growth and division (24 h p.i.; Fig. 2B and C).
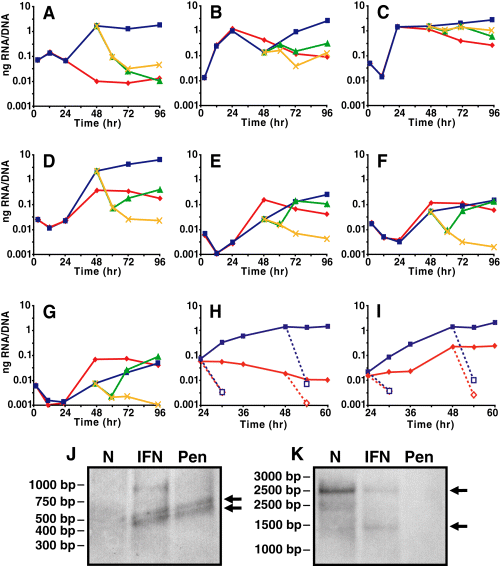
Detection of transcripts for various genes during IFNγ-treatment of Cpn-infected HEp-2 cells. Cells were infected and treated with or without IFNγ at the time of infection. A subset of samples was reactivated at 48 h p.i. in the presence or absence of penicillin. Quantitative RT-PCR normalized for genomic content for euo (A), groEL_1 (B), ompA (C), omcB (D), hctA (E), hctB (F), lcrH_1 (G). euo and omcB transcript levels after addition of rifampicin (dashed lines) (H and I). Data are representative of at least two replicate experiments performed in triplicate. Standard deviations are less than 5% of the sample. Equal amounts of 48 h p.i. RNA were loaded onto Northern blots for euo (J) and omcB (K) and processed in the absence of normalizing. The relative genome values are IFNg = 1; Normal = 14.9; Pen = 2.8. IFNγ and penicillin were added at the time of infection. Normal ( ); IFNγ (▪); reactivated (
); IFNγ (▪); reactivated ( ); reactivated with penicillin (
); reactivated with penicillin (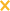 ).
).
Transcriptional profiles were very similar for all genes tested in untreated and IFNγ-treated comparisons until 24 h p.i. Between 24 h and 48 h p.i. in the IFNγ-treated samples, euo was highly upregulated (Fig. 2A) as has been published for Ctr (Belland et al., 2003a). Transcripts for the 60 kDa heat-shock chaperone encoded by groEL_1 decreased between 24 and 48 h p.i. but increased after this time (Fig. 2B); ompA transcripts remained elevated during IFNγ treatment (Fig. 2C). We anticipated that late gene transcription would be downregulated during IFNγ treatment because of the block in EB maturation. Surprisingly, the late-stage genes we analysed were upregulated between 24 h and 48 h p.i. during IFNγ treatment (Fig. 2D–G). Previous reports showed that late gene transcription was inhibited during IFNγ-mediated persistence (Mathews et al., 2001; Belland et al., 2003a; Goellner et al., 2006; Polkinghorne et al., 2006), which was expected as late proteins (histones, cysteine-rich outer envelope proteins) are not present in persistent forms. However, the transcription data in previous studies compared fold-differences between IFNγ-treated and untreated (i.e. a cross-sectional analysis) and were normalized to rRNA rather than DNA. Indeed, we found that the general induction of late gene transcription in C. pneumoniae in the presence of IFNγ was masked when data were normalized to 16S rRNA and analysed cross-sectionally (data not shown). In addition, we re-examined transcription in C. trachomatis and confirmed that late gene transcription was induced in the presence of IFNγ (see Fig. S1) when normalized to DNA. Therefore, the increase in late gene transcription that we observed was not only unexpected but also indicative of atypical regulatory systems.
In all reactivated samples, late gene transcript levels transiently returned to basal levels at 12 h post reactivation (60 h p.i.) before increasing again concomitantly with RB-to-EB differentiation. This increase in late gene transcription after removal of IFNγ could be attributed to residual effects of IFNγ host cell activation that would maintain the tryptophan limiting environment within the cell. However, samples reactivated in the presence of penicillin rapidly returned to 24 h p.i. transcription levels and remained low, verifying that the increase in late gene transcription after reactivation (without penicillin) was a result of the conversion of RBs to EBs and not a ‘return’ to the IFNγ-mediated profile.
To determine if the increase in transcription seen during IFNγ-mediated persistence was caused by an increase in transcript stability (due to decreased RNA degradation) rather than an increase in transcription initiation; rifampicin, a prokaryotic RNA polymerase inhibitor, was added to untreated and IFNγ-treated cultures at 24 h or 48 h p.i., and transcript levels were measured by qRT-PCR. Addition of rifampicin resulted in a rapid decrease in the amount of transcripts detected for euo and omcB (Fig. 2H and I) at both time points, supporting our hypothesis that the increase in transcript levels in the IFNγ-treated samples was due to increased transcription initiation and not to decreased RNA degradation.
To validate our qRT-PCR data, we performed Northern blots on equal amounts of RNA from 48 h p.i. to detect transcripts for euo and omcB in the absence of amplification or normalization. We detected transcripts for euo in the IFNγ- and penicillin-treated samples but not untreated controls whereas transcripts for omcB were detected in untreated and IFNγ-treated samples but not penicillin-treated samples (Fig. 2J and K, indicated by arrows). Detection of these transcripts in the IFNγ-treated samples is strongly indicative that these genes are transcribed during IFNγ-mediated persistence. The overall intensity of the omcB band was less in the IFNγ-treated sample because, in an equal amount of RNA, there are fewer genome equivalents in the IFNγ-treated compared with untreated samples (see Fig. 1). If our normalization methods were inappropriate, then we would not have been able to detect omcB transcripts.
Late gene expression is not activated during penicillin-induced persistence
We monitored selected gene transcription during penicillin treatment to establish if upregulated late gene expression is a general feature of persistence or an unusual characteristic of IFNγ-induced persistence. This is the first study to analyse chlamydial gene expression patterns in the presence of penicillin. Infected cells were treated with penicillin at the time of infection, and chlamydial DNA and transcript levels were quantified. Penicillin treatment prevented IFU recovery (data not shown), whereas chlamydial DNA levels increased over the course of the experiment (Fig. 3A), suggesting that DNA replication continued during penicillin-mediated persistence. 16S rRNA transcription remained constant after 24 h p.i. (Fig. 3B), and euo and late gene transcription was not upregulated after 24 h p.i. (Fig. 3C–E), in contrast to results with IFNγ treatment. These results indicated that: (i) late gene transcription was upregulated between 24 h and 48 h p.i. in IFNγ-mediated but not penicillin-mediated persistence; and (ii) penicillin treatment induced a transcriptional profile that was indistinguishable from an RB.
Quantitative PCR for genomic content and qRT-PCR for various genes during penicillin treatment of Cpn-infected HEp-2 cells. Cells were infected and treated with or without penicillin at the time of infection. Genomes (A), 16S rRNA (B), euo (C), omcB (D), hctB (E). Data are representative of at least two replicate experiments performed in triplicate. Standard deviations are less than 5% of the sample. Normal ( ); penicillin (▪).
); penicillin (▪).
Microarray analysis of C. pneumoniae grown in IFNγ-activated host cells indicates that transcription is globally upregulated
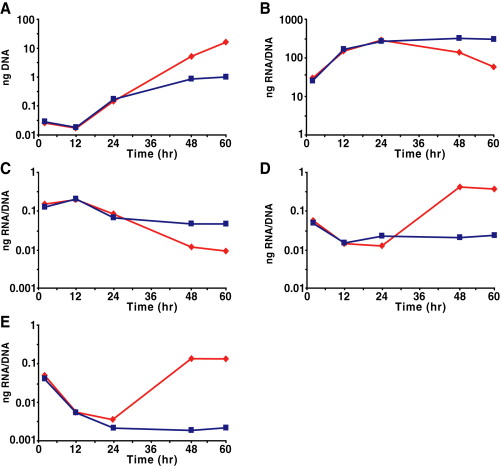
Microarray analysis was performed to validate our findings that late gene expression was activated during IFNγ-mediated persistence. We used a custom-designed Affymetrix gene chip with probe sequences tiled across every ORF. In addition, intergenic regions were detected using sequential 25mers. Late genes were characterized as those upregulated at least twofold between 24 h and 48 h p.i. during a normal developmental cycle (Fahr et al., 1995; Belland et al., 2003b). A selected set of these genes is listed in Table 1. When a comparison was made between 24 h and 48 h p.i. in the presence of IFNγ, late genes were found to be upregulated, confirming our qRT-PCR results. We analysed global transcription levels [ORFs and intergenic regions (IGRs)] by quantifying mean fluorescence intensity (MFI) of identically prepared samples normalized for genomic content. We found that, in the absence of IFNγ treatment, the MFI of ORFs decreased by 1.87-fold between 24 h and 48 h p.i. as RBs differentiated to EBs. More strikingly, we noticed a significant increase (6.28-fold upregulation for ORFs) in MFI over the same time frame during IFNγ treatment that returned to normal levels after reactivation (Fig. 4A and B), suggesting that transcription was globally upregulated for persistent cultures.
| Gene ID | Annotation | 48 h N array value | 48 h N qRT-PCR | 48 h IFN array value | 48 h IFN array SD | 48 h IFN qRT-PCR |
|---|---|---|---|---|---|---|
| Cpn0013 | pmpG-11 | 12.1 | 31.0 | 5.88 | ||
| Cpn0333 | ltuB | 3.72 | 5.42 | 4.37 | ||
| Cpn0378 | sucA | 7.94 | 12.1 | 11.7 | ||
| Cpn0384 | hctB | 4.46 | 31.2 | 5.66 | 3.38 | 17.1 |
| Cpn0445 | pmpG-7 | 8.74 | 3.14 | 0.07 | ||
| Cpn0557 | omcB | 4.93 | 17.0 | 83.3 | 35.8 | 100 |
| Cpn0558 | omcA | 9.45 | 103 | 37.5 | ||
| Cpn0607 | glgC | 15.4 | 30.8 | 5.92 | ||
| Cpn0670 | rsbW | 2.13 | 3.99 | 0.35 | ||
| Cpn0811 | lcrH_1 | 8.50 | 56.9 | 8.45 | 4.61 | 5.34 |
| Cpn0886 | hctA | 58.8 | 57.9 | 10.6 | 1.63 | 8.52 |
| Cpn0933 | 4.12 | 60.4 | 15.5 | |||
| Cpn1057 | yyaL | 9.21 | 11.3 | 3.59 | ||
| CpA0002 | 9.37 | 67.7 | 7.31 | |||
| CpA0004 | 10.6 | 30.6 | 0.10 |
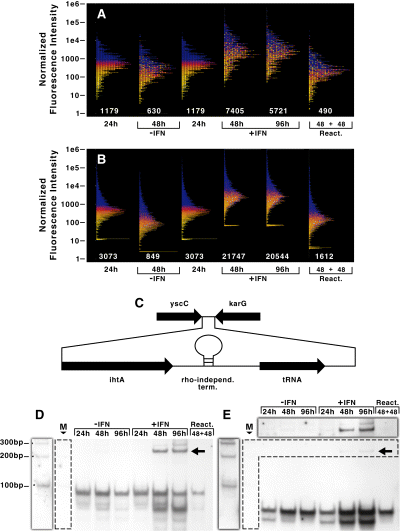
Microarray and Northern blot analysis of Cpn transcription during IFNγ-mediated persistence. Purified RNA samples were processed for microarray analysis as described in Experimental procedures. The normalized fluorescence intensity values for the complete genome encoding open reading frames (A) and intergenic regions (B) were plotted on a logarithmic scale. Numbers below each plot are the mean fluorescence intensity for the indicated sample. The colouring scheme is set according to expression levels at 24 h p.i. and are maintained within the other samples. Transcripts from intergenic regions were detected during IFNγ-treatment of infected HEp-2 cells.C. Schematic representation (not to scale) of the intergenic region in C. pneumoniae containing ihtA and threonyl-tRNA. Equal genomic equivalents of DNased total RNA were electrophoresed, transferred to nitrocellulose and probed for the indicated transcripts.D and E. D. Northern blot detecting ihtA transcripts during IFNγ-treatment of Cpn-infected HEp-2 cells. E. Northern blot detecting threonyl-tRNA transcripts during IFNγ-treatment of Cpn-infected HEp-2 cells. Shadowed regions outside of the blot correspond to a longer exposure of the regions bounded by the dashed line. M, molecular weight markers.
To verify that the global upregulation of transcription within intergenic regions applied to non-coding RNAs as well as the leader sequences of transcripts, we analysed the expression profile for one intergenic region by Northern blots. Recently, a small non-coding RNA, IhtA, was identified in C. trachomatis (Ctr) within an intergenic region and was postulated as a negative regulator of translation of HctA (Grieshaber et al., 2006). The C. pneumoniae (Cpn) ihtA was identified by homology within an intergenic region that also encodes a threonyl-tRNA and is bounded by two convergent ORFs (Fig. 4C). Between these non-coding RNAs is a predicted rho-independent terminator. Northern blots directed against Cpn ihtA and threonyl-tRNA were performed over a time-course for both IFNγ-treated and untreated cultures. In the untreated samples, IhtA and the tRNA were detected at all times but expression appeared to peak at 48 h p.i. (Fig. 4D and E). During IFNγ treatment, expression of both IhtA and the tRNA increased at 48 h p.i. and remained elevated (Fig. 4D and E), thus confirming the upregulation of IGRs detected by microarray analysis. After reactivation, ihtA and tRNAthr expression returned to control levels. Interestingly, at later time points during IFNγ treatment (48 h and 96 h p.i.), a band corresponding in size to IhtA (approximately 90 bp) was detected in addition to a larger form (approximately 220 bp) (Fig. 4D, indicated by the arrow). Subsequent experiments identified the larger band as a transcriptional fusion of both IhtA and the downstream tRNA. No transcript could be detected using a probe directed against the region upstream of the predicted start site of ihtA (data not shown). The tRNA (approximately 70 bp) also appeared upregulated in the IFNγ-treated samples (Fig. 4E). The proportion of high molecular weight bands appears to differ between the two blots because the tRNA is expressed at such high levels that we had to underexpose the blot to be able to see the tRNA clearly. This confounds any attempts to compare the IhtA and tRNA blots directly. These data indicated that the upregulation of transcription applied to non-coding RNAs and may have affected the normal processing of this intergenic region.
Cpn protein synthesis is decreased during IFNγ-mediated persistence and is inconsistent with transcriptional data
We carried out immunofluorescence (IF) microscopy studies to determine whether the upregulation of specific transcripts during treatment with IFNγ resulted in an increase in the accumulation of the corresponding proteins. We were particularly interested in the early-stage EUO and late-stage OmcB proteins because of the greater than 10-fold upregulation in gene expression between 24 h and 48 h p.i., as measured by qRT-PCR (Fig. 2). IF microscopy, albeit only semi-quantitative, allows detection of proteins within individual chlamydiae regardless of the size or shape of the organisms and therefore permitted us to directly compare protein accumulation at different times post infection and under different growth conditions without the need for normalization (Fig. 5A).
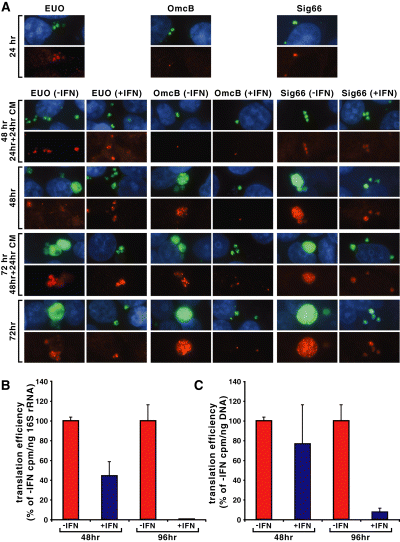
Analysis of protein content and translation during IFNγ-treatment of Cpn-infected HEp-2 cells. Indirect immunofluorescence of infected cells fixed in methanol and stained with primary antibody against the indicated antigen and secondary antibody goat anti-rabbit Alexa594 (red). EUO and OmcB panels were also stained with primary antibody against Cpn MOMP whereas Sig66 panels were stained with primary antibody against Hsp60-1; secondary antibody was goat anti-mouse Alexa488 (green). Host cell nuclei were visualized with Hoechst 33342. Some cultures were treated with the prokaryotic protein synthesis inhibitor chloramphenicol (CM) 24 h prior to fixation (A). Intrinsic pulse labelling with 35S-cys/met of infected cultures during IFNγ-treatment of Cpn-infected HEp-2 cells. Translation efficiency was defined as incorporated cpm per ng of 16S rRNA (B) or cpm per ng of genomic DNA (C) and the values for the untreated samples were arbitrarily set at 100%. Data shown are the average of three replicate experiments.
For detection of EUO and OmcB proteins, infected cells were reacted with rabbit polyclonal antisera directed against either EUO or OmcB, then secondarily with a fluorescence-tagged goat anti-rabbit immunoglobulin (red). To visualize chlamydiae within inclusions regardless of the presence of these proteins, the infected cells were also reacted with mouse monoclonal anti-MOMP (present in EBs, RBs and abnormal bodies) and a fluorescence-tagged goat anti-mouse secondary immunoglobulin (green). As anticipated, EUO was detected in untreated infections at 24 and 48 h p.i., but the fluorescence intensity faded considerably by 72 h p.i. EUO was also detected in abnormal forms in small inclusions in IFNγ-treated infected cultures at 24–72 h p.i. (24 h p.i. not shown); however, the intensity of the staining of the abnormal bodies in the inclusions was no greater than that of RBs in the control cultures. EUO antigen appears to be unstable by 72 h p.i. in the presence of IFNγ, as we noted some, but not all, inclusions at 72 h were completely devoid of EUO at this time. OmcB is present in EBs and consequently was detected at very early times post infection before conversion to the RB form (not shown). Interestingly, the OmcB antigen appears to be highly stable, as we still detected it at 24 h p.i. in the form of a single small EB-size dot even though two or three rounds of RB replication had occurred. By 48 h p.i., newly synthesized OmcB accumulated in the inclusion and OmcB continued to accumulate through 72 h p.i. in untreated infected cells. In IFNγ-treated infected cells, we detected OmcB only in the original infecting EB at all times post infection; that is, no new synthesis of OmcB had occurred. We also examined accumulation of the major sigma factor Sig66 protein (red fluorescence) and GroEL_1 protein (green) and found them to be present at all times post infection at approximately the same intensity in the presence or absence of IFNγ. We conclude from these studies that the upregulation of gene transcription in the presence of IFNγ does not result in a notable increase in protein synthesis, even in the case of the highly upregulated EUO and OmcB genes. Indeed, in the case of OmcB, there is no net accumulation of protein in the presence of IFNγ.
To determine whether or not the failure to detect increased protein accumulation in the presence of IFNγ was due to a decrease in stability of the proteins, the prokaryotic protein synthesis inhibitor chloramphenicol was added at 24 and 48 h p.i. and the cells were examined by IF microscopy 24 h later. We found the amount of protein present in all cases, with the exception of EUO at late times in the presence of IFNγ noted earlier, appeared to be the same after 24 h of chloramphenicol treatment as at the time of addition of the protein synthesis inhibitor. We conclude that OmcB, MOMP, Sig66 and GroEL_1 are relatively stable at all times both in the presence and the absence of IFNγ and that EUO is relatively stable through 48 h p.i.
We next investigated if chlamydial protein synthesis was generally reduced during IFNγ treatment. Untreated and IFNγ-treated infected and uninfected cells were labelled with 35S-cys/met in the presence of the eukaryotic protein synthesis inhibitor emetine and then harvested for analysis. Equivalent amounts of protein were precipitated in 10% TCA and collected on filters. Total cpm were measured and normalized to both 16S rRNA (Fig. 5B) and genome levels (Fig. 5C). Translational efficiency, as measured by the cpm per 16S rRNA equivalent, was more than twofold reduced (P < 0.0025; one-tailed t-test) at 48 h p.i. in IFNγ-treated cultures compared with untreated (Fig. 5B) whereas cpm per genome was not significantly different (P < 0.25) (Fig. 5C). More surprisingly, cpm per 16S rRNA (P < 0.005) or cpm per genome (P < 0.0025) at 96 h p.i. in IFNγ-treated cultures was barely above the eukaryotic background levels, indicating that translation was severely reduced in these cultures. Reactivating the IFNγ-treated cultures at 96 h p.i. led to the development of mature inclusions (data not shown), suggesting that the abrogation of translation was not due to chlamydial death. Pulse-chase samples for both IFNγ-treated and untreated cultures showed no significant decrease in cpm after the chase (data not shown), verifying that protein stability was maintained and corroborating our chloramphenicol IFA data. Together, these observations indicated that chlamydial proteins were highly stable and helped to explain how chlamydiae survive IFNγ-induced tryptophan limitation in the absence of efficient, nascent translation.
Discussion
Treatment of Chlamydia-infected human epithelial cell lines in vitro with IFNγ induces an aberrant growth state in these bacteria (Beatty et al., 1993), called persistence, by virtue of this cytokine's ability to induce a tryptophan-limiting environment within the cell (Pfefferkorn, 1984; Byrne et al., 1986). There is much interest in elucidating the mechanisms of chlamydial persistence induction and reactivation due to their importance for understanding immune-mediated chronic disease and for developing improved therapeutics. Consequently, the relevance of the IFNγ model of persistence in Chlamydia has made it a target of investigations, and many reports have analysed transcript and protein profiles during this state of stress (Shaw et al., 1999; Mathews et al., 2001; Belland et al., 2003a; Goellner et al., 2006; Mukhopadhyay et al., 2006; Polkinghorne et al., 2006).
We initiated studies to determine the gene expression patterns of C. pneumoniae to IFNγ-mediated tryptophan starvation with the rationale that this organism induces a persistent state by engaging a specific transcriptional response or regulon (Belland et al., 2003a). Previous studies measuring chlamydial transcription during IFNγ-mediated persistence relied on a direct comparison between the treated and untreated samples (i.e. cross-sectional) to acquire a fold up- or downregulated change in expression (Mathews et al., 2001; Belland et al., 2003a; Goellner et al., 2006; Polkinghorne et al., 2006). This method of analysis does not identify the transcriptional changes that occur longitudinally within treated samples (e.g. 24 h IFN versus 48 h IFN). At the completion of a normal developmental cycle, RBs differentiate to EBs. During IFNγ treatment, the RB grows and divides one or two times before becoming abnormal, and persistent forms do not differentiate to EBs. Consequently, earlier work compared transcription of persistent forms with that of differentiating RBs whereas a more appropriate comparison would be to monitor the changes that occur as the RB progresses to a persistent form. Therefore, we chose to investigate longitudinal transcriptional changes within each sample set and to determine the chlamydial response to IFNγ treatment.
Most previous studies used the 16S rRNA gene to normalize transcript levels, assuming that 16S rRNA transcripts per genome were equal under each condition examined. Our data demonstrate that 16S rRNA transcription, as measured on a per genome basis, peaks at mid-cycle (24 h p.i.) then declines during normal growth while it remains elevated for the duration of the experiment (96 h) during IFNγ-mediated persistent growth (1, 3). Therefore, we chose to normalize transcript data to genomic content rather than the more traditional 16S rRNA normalization procedure. This method may have broad applicability for prokaryotic transcriptional studies, particularly those involving bacterial developmental gene regulation or transitions from logarithmic growth to stationary phase.
By using these analytical methods, we have made three intriguing observations from our transcriptional and translational analyses over a time-course of IFNγ treatment and reactivation from persistence in Chlamydia. Firstly, ORF and intergenic region transcription were uniformly upregulated during IFNγ-mediated persistence (Fig. 4). Although the total biomass of RNA (and organism) in IFNγ-treated cultures was less than in the untreated cultures because of reduced growth and division (see Fig. 1), the number of transcripts per genome was uniformly elevated. This is an unusual response for a bacterium during a stress-induced state. Secondly, and in contrast to published reports (Mathews et al., 2001; Belland et al., 2003a; Slepenkin et al., 2003; Goellner et al., 2006; Polkinghorne et al., 2006), EB-associated (late) genes involved in the secondary differentiation from RBs to EBs were induced during IFNγ treatment, which is surprising given the blockage in EB maturation (Figs. 2 and S1, Table 1). This profile is not a generalized response to persistence because penicillin treatment fails to elicit the same profile (Fig. 3); indeed, late genes are not activated during penicillin treatment. These data indicate that chlamydial developmental gene regulation does not function properly during IFNγ-mediated persistence. Finally, chlamydial protein synthesis was significantly reduced during IFNγ treatment, indicating an uncoupling of transcription from translation (Fig. 5). This finding is contrary to the traditional view that transcription and translation are linked in bacteria.
In the absence of efficient translation, protein stability may become important in maintaining viability of persistent organisms. Indeed, our IFA data of cultures treated with a protein synthesis inhibitor showed that the chlamydial protein staining patterns were maintained after addition of the inhibitor (Fig. 5A). Pulse-chase experiments further validated this observation as there was no decrease in the incorporated radioactivity during the chase (data not shown). Therefore, chlamydial proteins are quite stable and may be important in maintaining viability of IFNγ-induced persistent forms.
IFNγ activation of human cell lines results in a tryptophan-limiting environment within the host cell, which will starve the tryptophan-auxotrophic Chlamydia. In most bacteria starvation for amino acids triggers the stringent response, a transcriptional programme evolved to overcome amino acid starvation. When an uncharged tRNA binds in the ribosome, the ribosome-associated RelA protein is activated to synthesize ppGpp (Chatterji and Ojha, 2001), which acts as a global regulator of transcription by modulating transcription complexes at promoters (Magnusson et al., 2005). The stringent response serves to stop the synthesis of stable RNA species, such as rRNA and tRNA, to increase protein degradation pathways, and to increase synthesis of enzymes involved in amino acid biosynthesis (Chatterji and Ojha, 2001). Collectively, these responses serve to overcome the amino acid limitation. SpoT is a cytosolic bifunctional enzyme with ppGpp synthase and hydrolase activity that helps control the levels of ppGpp. Chlamydia and other obligate intracellular pathogens are predicted to lack a stringent response as they have no obvious homologues of RelA and SpoT and do not synthesize ppGpp (Mittenhuber, 2001). The loss of these genes has likely occurred through reductive evolution as a means for adapting to obligate intracellular growth. The absence of relA and spoT homologues, the inability to synthesize ppGpp and the upregulation of stable RNA (1, 4 and S1) indicate Chlamydia do not engage a stringent response during IFNγ-mediated tryptophan starvation.
Transcriptional upregulation in the absence of translation is unusual and, in the absence of a stringent response, may represent a default pathway for organisms under amino acid limitation. One possible explanation consistent with our data is that tryptophan starvation may prevent ribosomes from synthesizing proteins that maintain developmental transcriptional control of the organism, and thus the global upregulation more accurately reflects a lack of regulation. This explanation invokes an inability to regulate transcription as the cause of global upregulation. Our observations suggest Chlamydia have lost the mechanisms required for regulating transcription during amino acid starvation, and this may be a general phenomenon among bacteria that lack stringent responses.
A fundamental question is why Chlamydia, in the absence of a stringent response, evolved to globally upregulate transcription during IFNγ-mediated tryptophan starvation. The answer to this is unknown, but it is possible that transcriptional upregulation could confer an advantage to the organism during reactivation to productive growth. The abundance of transcripts might allow for rapid translation of proteins once tryptophan becomes available. Therefore, the upregulation of transcription may not be evolved exclusively to enact specific changes in the organism to respond directly to tryptophan starvation but also to aid in the return to normal growth with quick production of EBs. The present data allow us to rule out potential mechanisms for our observations during IFNγ-mediated persistence. Firstly, the global increase in transcripts is not due to decreased RNA degradation but reflects increased initiation. Secondly, protein synthesis decreases because of inefficient translation, resulting in an apparent disconnect between transcription and translation. These observations will further our understanding of chlamydial persistence and may facilitate studies in other organisms lacking stringent responses.
Experimental procedures
Organisms and cell culture
Chlamydia pneumoniae AR39 was originally plaque purified according to the scheme of Gieffers et al. (2002), and subsequent passages were grown in HEp-2 cells at 35°C with 7% CO2. EBs were harvested from infected cell cultures, purified by discontinuous density gradient centrifugation in Renografin (Bracco Diagnostics, Princeton, NJ), and titerd for infectivity as measured by IFU. Similarly, EBs were harvested from C. trachomatis serovar D grown at 37°C with 7% CO2. HEp-2 cells were incubated at 37°C with 5% CO2 in IMDM (Cambrex/Biowhittaker, Walkersville, MD) supplemented with 10% fetal bovine serum (Hyclone, Logan, UT), 2 mM l-glutamine and 10 μg ml−1 gentamicin (Gibco/Invitrogen, Carlsbad, CA). All chemicals were from Sigma Chemical (St Louis, MO) unless otherwise noted.
Preparation of cells for infection
HEp-2 cells were plated in six-well culture plates at a density of 1.2 × 106 cells per well. In a subset of wells, cells were plated onto glass coverslips for immunofluorescence microscopy. Approximately 18 h later, confluent cell monolayers were rinsed with Hanks' balanced salt solution (HBSS; Gibco), and 2 ml of inoculum containing 2.6 × 106 Cpn IFUs in SPG (0.25 M sucrose, 10 mM sodium phosphate and 5 mM l-glutamic acid) was added to each well. Infected cell cultures were centrifuged in an RC-3B Sorvall centrifuge for 1 h at 30°C, 400 g then rocked for 30 min at 35°C without CO2. The inoculum was aspirated, and fresh supplemented IMDM with or without 0.5 ng ml−1 recombinant human IFNγ (Biosource, Camarillo, CA) or 5 μg ml−1 penicillin G was added to each well. Infected cultures were incubated at 35°C with 7% CO2. For Ctr HEp-2 cells were treated with or without 0.5 ng ml−1 IFNγ 24 h prior to infection. At the time of infection, 6 × 106 IFUs in 10 μl were added directly to each well, and infected cells were incubated at 37°C with 7% CO2. For reactivated cultures, IFNγ was aspirated from a subset of infected samples, cells were washed three times with HBSS, and fresh IMDM containing 64 μg ml−1 tryptophan with or without 20 μg ml−1 penicillin G was added to the cells. For all samples addition of inoculum to wells marks the time of infection (t = 0 h). The dose of IFNγ used was determined empirically by titrating the cytokine on infected cell monolayers and assessing EB production. A dose was used that gave minimal recovery at 48 h p.i. for Cpn or 24 h p.i. for Ctr but maximal recovery after 48 h of reactivation for Cpn or 24 h for Ctr (total 96 h or 48 h p.i., respectively; data not shown).
Quantification of IFUs from infected cell cultures
Medium was aspirated from the treatment samples of infected cells at the indicated times post infection, and 1 ml of SPG was added to each well. Cells were scraped and collected from each well into a 2 ml microfuge tube with three glass beads. Samples were vortexed for 45 s and frozen at −80°C. Samples were titred for infectivity on fresh cell layers as described earlier to quantify the number of IFU per well.
Preparation of cells for electron microscopy
At 48 and 96 h p.i., cells from infected cultures were collected and centrifuged to a pellet for 5 min at 1500 r.p.m. Cell pellets were resuspended in 1% EM-grade glutaraldehyde (Fluka/Sigma) diluted in PBS, transferred to a microfuge tube, and centrifuged to a pellet at 3000 r.p.m. for 5 min. Cells were subsequently processed for EM as described elsewhere (Byrne et al., 2001). Briefly, cells were washed three times with PBS then fixed for 1 h at room temperature in 1% osmium tetroxide diluted in PBS. Samples were dehydrated with a graded series of ethanol and embedded in Spurr's resin (Electron Microscopy Sciences, Ft. Washington, PA). In this study 70–80 nm sections were cut, stained with uranyl acetate and lead citrate, and viewed on a Zeiss transmission electron microscope. Electron microscopy was performed by Wandy Beatty at Washington University St Louis.
Isolation and purification of nucleic acids
At the indicated times post infection, medium from infected cultures was aspirated, and cells were washed with HBSS and subsequently lysed with Trizol reagent (Invitrogen). 0.2 volume of chloroform was added to the Trizol lysate, mixed thoroughly, and centrifuged for 5 min to separate the phases. The upper aqueous phase containing RNA was collected into fresh tubes and precipitated with an equal volume of isopropanol. RNA samples were further purified using Oligotex to remove poly-adenylated host mRNA according to the manufacturer's instructions (Qiagen, Valencia, CA) and rigorously DNased using Turbo DNAfree (Ambion). From duplicate infected samples, cells were collected, centrifuged to a pellet, and resuspended in 500 μl of phosphate buffered saline (PBS; Gibco). DNA was isolated from these samples using a DNeasy kit according to the manufacturer's instructions (Qiagen). All nucleic acid concentrations were quantified (A260) using a Nanodrop spectrophotometer, and appropriate aliquots were made and stored at −80°C for RNA and −30°C for DNA.
Quantification of genomes from infected cells using qPCR
TaqMan primer/probe sets for indicated genes were designed using the Primer Express software (Applied Biosystems, Foster City, CA) and tested against C. pneumoniae or C. trachomatis chromosomal DNA (Table S1). qPCR was performed against Cpn0423 or Ctr groEL_3 using 150 ng of total DNA per well from each sample as previously reported with the TaqMan Universal PCR Master Mix (Applied Biosystems) and the ABI Prism 7000 Sequence Detection System (Applied Biosystems) (Ouellette et al., 2005). Human genomes were quantified using the TaqMan β-actin Control Reagents (Applied Biosystems) from the same DNA aliquots. The amount of DNA was determined for each sample by converting mean critical threshold values (typically between 20 and 30 Ct with 150 ng sample size) to ng of DNA using standard curves for each primer/probe set generated against chromosomal DNA from their respective organism as described elsewhere. Chlamydial DNA was normalized to human DNA as a loading control. Table S1 shows a list of all primer/probe sequences used.
Quantification of transcripts from infected cells using qRT-PCR
Transcripts were quantified from 20 ng of purified RNA at the indicated time points using the TaqMan One-Step RT-PCR Master Mix Reagents (Applied Biosystems) and the ABI Prism 7000 Sequence Detection System. The amount of transcripts was quantified as above for DNA. Transcript levels were normalized to corrected chlamydial DNA to give relative amounts of transcripts per DNA. Transcripts were normalized to genomes as opposed to 16S rRNA because we have found that the levels of 16S rRNA per genome do not accurately reflect the number of organisms during the course of infection (see Results), which is a prerequisite for using it as a control.
Preparation of samples for microarray analysis
DNased total RNA that had been treated with Oligotex to remove host poly-adenylated mRNA was further purified for microarray analysis using the MicrobEnrich and MicrobExpress kits (Ambion) according to the manufacturer's instructions. Purified RNA was prepared for hybridization to the Affymetrix gene chip by synthesizing antisense cDNA and endlabelling it according to the Affymetrix protocol for prokaryotic samples described in GeneChip Expression Analysis Technical manual. Hybridization occurred for 16 h at 45°C in an Affymetrix Hybridization Oven 640 at 60 r.p.m. Samples were washed and stained on the Affymetrix Fluidics Station 450 using an anti-streptavidin antibody (Vector Laboratories) and R-streptavidin phycoerythrin (Molecular Probes).
Analysis of microarray data
Gene chips were scanned using the Affymetrix GeneChip Scanner 3000. Fluorescence data were scaled using GCOS1.1 (Affymetrix) to a mean intensity of 1000. Data were corrected for genomic content using the measurements obtained by qPCR from the DNA samples. Data imported into GeneSpring were normalized using the ‘per chip: normalize to constant value’ normalization scheme. Distribution plots were generated by setting the parameters to display as non-continuous and using the line graph visualization feature in GeneSpring.
Northern blots
For euo and omcB (Fig. 2), equal amounts of DNased total RNA were electrophoresed on a 1.5% or 1%, respectively, agarose gel and transferred to nitrocellulose. For IhtA and threonyl-tRNA (Fig. 4), equal genomic equivalents of DNased total RNA for each time point analysed were electrophoresed on a 10% Urea-PAGE gel with TBE buffer and subsequently transferred to a nitrocellulose blot (Bio-Rad Laboratories, Hercules, CA). Biotinylated Northern probes were generated against the designated sequences using primers listed in Table S1 and the Megascript kit (Ambion). RNA sequences were probed following the NorthernMax protocol and detected with the BrightStar BioDetect kit (Ambion). Blots were then exposed to film and developed.
Preparation of cells for immunofluorescence
At various times post infection, cells plated on coverslips were washed with PBS, fixed with methanol for 10 min, and carefully washed three times with PBS plus 0.025% sodium azide. For a subset of analyses, infected cells were treated with 100 μg ml−1 chloramphenicol and further incubated for 24 h prior to fixation. Infected Cpn cultures were stained with primary mouse monoclonal antibody (Ab) GZD1E8 against Cpn MOMP (Wolf et al., 2001) or A57-B9 against Hsp60_1 (Beatty et al., 1993) and secondary Ab goat anti-mouse Alexa488 (Molecular Probes/Invitrogen). Polyclonal rabbit primary antibodies against EUO, OmcB and Sig66 were also used in conjunction with a secondary goat anti-rabbit Alexa594 Ab (Molecular Probes/Invitrogen). Host cell nuclei were visualized with Hoechst 33342. Immunofluorescence was viewed on an Axioplan 2 microscope (Carl Zeiss, Germany).
Intrinsic pulse/chase labelling
At 48 h or 96 h p.i., untreated or IFNγ-treated infected cells were incubated for 45 min in custom-made IMDM lacking cysteine and methionine (cmIMDM) plus 20 μg ml−1 of the eukaryotic protein synthesis inhibitor emetine. Following this starvation, 120 μCi of 35S-cys/met Pro-mix (Amersham Biosciences, Piscataway, NJ) in cmIMDM plus 20 μg ml−1 emetine was added to cell cultures for 45 min. Cells were washed three times in cmIMDM containing 3× cys and 3× met. Cells were either collected, centrifuged, resuspended in 250 μl of PBS, and frozen for the pulse-labelling experiments or incubated in fresh IMDM with or without tryptophan for the chase experiments and collected at subsequent times. Equal amounts of protein were precipitated in 10% TCA, collected on filters, and added to 5 ml of Bio-Safe II scintillation fluid (Research Products International Corporation, Mount Prospect, IL). Total cpm was collected from samples in a scintillation counter (Beckman Coulter, Fullerton, CA).
Acknowledgements
These data were presented in part at the 105th General Meeting of the American Society of Microbiology, 5–9 June, 2005 in Atlanta, GA. The authors are grateful to Harlan Caldwell for providing antibodies to MOMP and for encouraging comments. The authors would like to thank Dr Jon Katze for critical review of the manuscript and Tim Higgins for his hard work with figures. This work was funded in part by NIH grants AI30040 to G.I.B., HL71735 to G.I.B. and R.J.B., and AI19570 to T.P.H.