Mycobacterial FurA is a negative regulator of catalase–peroxidase gene katG
Abstract
In several bacteria, the catalase–peroxidase gene katG is under positive control by oxyR, a transcriptional regulator of the peroxide stress response. The Mycobacterium tuberculosis genome also contains sequences corresponding to oxyR, but this gene has been inactivated in the tubercle bacillus because of the presence of multiple mutations and deletions. Thus, M. tuberculosis katG and possibly other parts of the oxidative stress response in this organism are either not regulated or are controlled by a factor different from OxyR. The mycobacterial FurA is a homologue of the ferric uptake regulator Fur and is encoded by a gene located immediately upstream of katG. Here, we examine the possibility that FurA regulates katG expression. Inactivation of furA on the Mycobacterium smegmatis chromosome, a mycobacterial species that also lacks an oxyR homologue, resulted in derepression of katG, concomitant with increased resistance of the furA mutant to H2O2. In addition, M. smegmatis furA::Kmr was more sensitive to the front-line antituberculosis agent isonicotinic acid hydrazide (INH) compared with the parental furA+ strain. The phenotypic manifestations were specific, as the mutant strain did not show altered sensitivity to organic peroxides, and both H2O2 and INH susceptibility profiles were complemented by the wild-type furA+ gene. We conclude that FurA is a second regulator of oxidative stress response in mycobacteria and that it negatively controls katG. In species lacking a functional oxyR, such as M. tuberculosis and M. smegmatis, FurA appears to be a dominant regulator affecting mycobacterial physiology and intracellular survival.
Introduction
Mycobacterium tuberculosis is a facultative intracellular pathogen capable of surviving and persisting in phagocytic cells (Armstrong and Hart, 1971; Dannenberg et al., 1994). The ability to resist killing by reactive oxygen (Chan et al., 1991; Yuan et al., 1995; Manca et al., 1999) and nitrogen intermediates (Yu et al., 1999), prevent phagosomal maturation into the phagolysosome (Armstrong et al., 1971; Clemens and Horwitz, 1995; Russell, 1995; Deretic and Fratti, 1999) and avoid detection and elimination by the host's immune system (Pancholi et al., 1993; Stenger et al., 1998; Mustafa et al., 1999) may contribute to the overall potency of this pathogen. In this context, genes and factors involved in protection against oxidative stress are likely to play a role in detoxification of reactive oxygen and nitrogen intermediates encountered upon entry and during residence in infected macrophages. Specifically, the oxidative stress response genes encoding the catalase-peroxidase (katG) and the catalytic subunit of alkyl hydroperoxidase reductase (ahpC) have been implicated in intracellular survival and persistence of pathogenic mycobacteria in the host (Middlebrook and Cohn, 1953; Morse et al., 1954; Mitchison et al., 1963; Wilson et al., 1995; 1998; Heym et al., 1997; Chen et al., 1998; Li et al., 1998; Manca et al., 1999; Cooper et al., 2000). In parallel to the potential role in pathogenicity, these and additional determinants have been shown to participate in acquired resistance (Zhang et al., 1992; Banerjee et al., 1994; Heym et al., 1995; Musser, 1995; Rouse and Morris, 1995; Mdluli et al., 1998) and innate susceptibility to isonicotinic acid hydrazide (INH) (Deretic et al., 1996; Zhang et al., 1996).
Understanding factors that contribute to the regulation of oxidative stress response should also aid in the dissection of host–pathogen interactions associated with mycobacterial diseases. In enteric bacteria, the oxidative stress response is mediated by the regulated expression of katG and ahpC in addition to other factors (Storz and Imlay, 1999). Both genes, as well as several others (Christman et al., 1985; Tartaglia et al., 1989; Altuvia et al., 1994), are under the positive regulation of oxyR, a central transcriptional regulator of the peroxide stress response (Christman et al., 1989; Storz and Altuvia, 1994). In M. tuberculosis and other members of the M. tuberculosis complex (Mycobacterium bovis, Mycobacterium africanum and Mycobacterium microti), the orthologue of oxyR has been rendered inactive via multiple mutations (Fig. 1A) (Deretic et al., 1995; Sherman et al., 1995). Consequently, only limited induction of katG and ahpC in M. tuberculosis can be observed upon exposure to peroxides, possibly contributing to the exquisite sensitivity of M. tuberculosis to INH (Deretic et al., 1996; Zhang et al., 1996). In contrast, Mycobacterium leprae, an organism not sensitive to INH, possesses complete oxyR and ahpC genes, but has multiple lesions in the katG gene (Eiglmeier et al., 1997; Nakata et al., 1997). Thus, the absence of functional components of the oxidative stress response appears to be a common theme in pathogenic mycobacteria (Deretic et al., 1997), albeit such phenomena appear counterintuitive in the context of intracellular survival in infected macrophages (Chan and Kaufmann, 1994). In addition to the partial elimination of elements of the oxidative stress response in the two major human mycobacterial pathogens, M. tuberculosis and M. leprae, several non-pathogenic mycobacterial species also differ from the enterobacterial paradigm of oxidative stress response. For example, the fast-growing species Mycobacterium smegmatis displays induced expression of at least nine proteins (Dhandayuthapani et al., 1996) in response to peroxide stress, including katG (Sherman et al., 1995) and ahpC (Sherman et al., 1995; Dhandayuthapani et al., 1996), although it lacks an oxyR equivalent (Fig. 1A). Furthermore, in Mycobacterium marinum, a pathogenic non-tuberculous species (Falkinham, 1996) that is relatively closely related to M. tuberculosis (Rogall et al., 1990), oxyR does not regulate expression of katG although it controls ahpC (Pagan-Ramos et al., 1998). Thus, it seems plausible that genes and mechanisms different from those previously identified in Enterobacteriaceae may control aspects of oxidative stress response in mycobacteria.
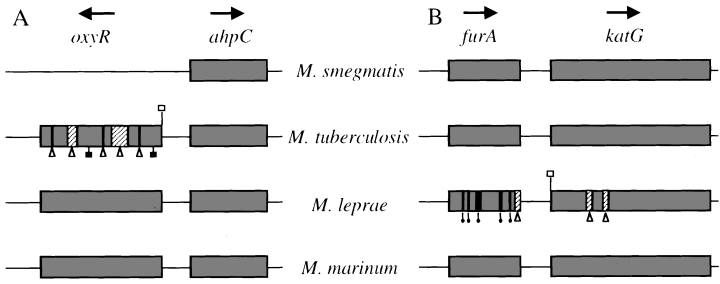
Genetic organization of the ahpC–oxyR (A) and furA–katG (B) regions in mycobacteria. Arrows, direction of transcription; open triangles below hatched segments, large deletions; triangles below lines, frameshift mutations; filled balloons, frameshift insertions; open squares, mutations in the start codon; closed squares, nonsense mutations.
Apart from oxyR-dependent regulation of oxidative stress response, many organisms couple the expression of oxidative stress genes with iron metabolism, principally via the ferric uptake regulator Fur. Fur or Fur homologues regulate genes induced in response to oxidative stress, including sodA (Niederhoffer et al., 1990; Tardat and Touati, 1993) and sodB (Niederhoffer et al., 1990; Dubrac et al., 2000) encoding Mn2+ and Fe2+ superoxide dismutases (SOD), the 8-hydroxyguanine endonuclease gene (Lee et al., 1998), catalase and peroxidase genes (Hassett et al., 1997; Bsat et al., 1998; van Vliet et al., 1998; 1999; Baillon et al., 1999), alkyl hydroperoxidase genes (Bsat et al., 1998; van Vliet et al., 1998; 1999), the soxRS genes (Zheng et al., 1999) and even the oxyR gene (Zheng et al., 1999). Mycobacteria also appear to couple expression of oxidative stress genes with iron metabolism. For example, inactivation of ideR, encoding an orthologue of the iron-responsive Cornybacterium diphtheriae repressor DtxR, renders M. smegmatis more sensitive to H2O2 as a result of decreased KatG and Mn2+ SOD activities (Dussurget et al., 1996).
The presence of a fur-like gene, furA, immediately upstream of katG in several mycobacterial species including M. tuberculosis, M. leprae and M. marinum (Fig. 1B) led us to propose that a subset of oxidative stress response genes may be regulated by FurA (Deretic et al., 1997; Pagan-Ramos et al., 1998). To investigate further the potential role of furA in the regulation of oxidative stress genes in mycobacteria, we characterized the furA gene of M. smegmatis and tested its regulatory role in the oxidative stress response. We present evidence demonstrating that furA negatively regulates katG expression. Inactivation of furA on the chromosome of M. smegmatis increased both the resistance of this organism to H2O2 and its susceptibility to INH. These observations have implications for the virulence of mycobacteria in the context of the peculiar evolutionary events that lead to a deregulation or partial elimination of oxidative stress response in pathogenic mycobacteria.
Results
The furA and katG genes are genetically linked in M. smegmatis and other mycobacteria
The furA gene is located immediately upstream of katG in M. tuberculosis, M. leprae and M. marinum (Deretic et al., 1997; Pagan-Ramos et al., 1998). To determine whether the furA and katG linkage is also conserved in M. smegmatis, we cloned and sequenced the region immediately upstream of katG in M. smegmatis mc2155. An open reading frame (ORF; GenBank accession number AF012631) encoding a polypeptide of 126 amino acids with high homology to FurA from other mycobacterial species was present immediately upstream of katG. M. smegmatis FurA was 65% identical to M. tuberculosis FurA and 57% identical to M. marinum FurA (data not shown). Additional polymerase chain reaction (PCR) analysis of diverse mycobacterial species using degenerate primers based on conserved regions of furA and katG also detected the furA–katG linkage in Mycobacterium avium, Mycobacterium aurum, Mycobacterium xenopi and Mycobacterium fortuitum, in addition to M. tuberculosis and M. marinum (Fig. 2A). The corresponding DNA fragments containing partial furA and katG genes were subjected to DNA sequencing and their identity confirmed (GenBank accession numbers AF092559, AF092558 and AF092560). Thus, the furA–katG linkage was found to be conserved in all six of the mycobacteria species analysed.
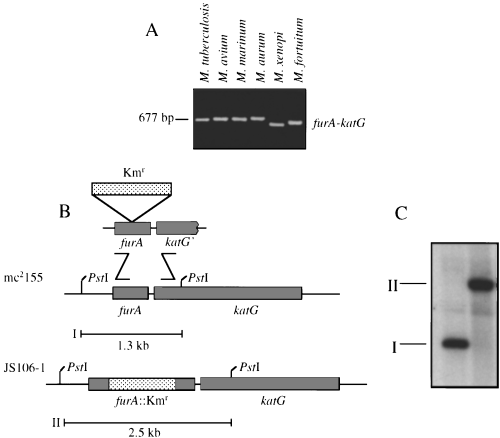
Conservation of furA–katG linkage in mycobacterial species and inactivation of the furA gene in M. smegmatis mc2155 via homologous recombination.
A. Two degenerate primers, FurAZooL and FurAZooR (see Experimental procedures), corresponding to conserved regions of furA and katG were used to amplify the corresponding regions from M. tuberculosis, M. avium, M. marinum, M. aurum, M. xenopi and M. fortuitum. The identity of PCR fragments as furA–katG was confirmed by sequencing.
B. Inactivation of the furA gene in M. smegmatis mc2155 via homologous recombination. The furA gene from M. smegmatis was disrupted by allelic exchange using a linear DNA fragment containing the furA gene with a Kmr cassette insertion.
C. Southern blot hybridization analysis using PstI-digested chromosomal DNA and a furA-specific radiolabelled probe. I, 1.3 kb PstI fragment from wild-type M. smegmatis mc2155; II, 2.5 kb PstI fragment from furA::Kmr isolate JS106-1 (see scheme in B).
Gene replacements with furA::Kmr in M. smegmatis and analysis of phenotypic effects on sensitivity to oxidants
The ubiquitous presence in mycobacteria of furA immediately upstream of the catalase–peroxidase gene katG led us to investigate whether the expression of oxidative stress response genes in M. smegmatis was regulated by furA. To this end, we inactivated furA on the chromosome of M. smegmatis mc2155. Using a linear DNA fragment carrying a disrupted copy of furA, furA::Kmr mutants were generated in M. smegmatis by gene replacement via homologous recombination (Fig. 2B). Southern blot analysis performed on one such recombinant, JS106-1, confirmed that a true gene replacement of furA+ with furA::Kmr had occurred (Fig. 2C). To determine whether inactivation of furA in M. smegmatis grown in 7H9 medium had phenotypic effects on oxidative stress, we examined the susceptibility of JS106-1 to peroxides (Table 1). Somewhat unexpectedly, JS106-1 (furA::Kmr) was significantly more resistant to H2O2 than the parental furA+ strain M. smegmatis mc2155 (Table 1, rows A and B; P = 0.0001, anova). The decreased sensitivity to H2O2 was a direct result of the furA::Kmr alteration, because complementation of the furA::Kmr mutation in JS106-1 by the introduction of pMsFurA carrying a wild-type furA+ gene (strain JS106-1 [pMsFurA]) reversed the increased resistance to H2O2 (Table 1, rows B and C; P = 0.0001, anova). When sensitivity to cumene hydroperoxide (CHP) was tested, no significant differences in susceptibility to this organic peroxide were observed between mc2155, JS106-1 and JS106-1 [pMsFurA] strains (Table 1, rows A–C). In contrast, an ahpC::Kmr mutant of M. smegmatis mc2155, VD1865-6, showed increased sensitivity to CHP (Table 1, row D; P = 0.0001, anova). These results are consistent with the interpretation that inactivation of furA derepresses a system specifically involved in detoxification of hydrogen peroxide but not organic peroxides.
| Row | Strainb | Genotype | H2O2 | CHP | INH |
|---|---|---|---|---|---|
| A | mc2155 | furA + | 31.3 ± 0.3 | 31.7 ± 0.3 | 20.3 ± 0.3 |
| B | JS106-1 | furA::Kmr | 26.7 ± 0.3 | 31.7 ± 0.3 | 50.3 ± 0.6 |
| C | JS106-1 [pMsFurA] | furA::Kmr[furA+] | 39.7 ± 0.3 | 31.3 ± 0.3 | 31.7 ± 0.3 |
| D | VD1865-6 | ahpC::Kmr | 35.0 ± 0.6 | 38.7 ± 0.3 | 36.3 ± 0.3 |
- a . Values represent the mean diameter ± SE (in mm) of zones of inhibition (experiments performed in triplicate) from strains grown in 7H9 medium. Filter disks (quarter-inch diameter) were soaked with 10 µl of 2% H 2O2, 2% CHP or 1 mg ml−1 INH. Results were read 3 days after plating.
- b . JS106-1, JS106-1 [pMsFurA] and VD1865-6 are isogenic derivatives of M. smegmatis mc 2155.
The furA gene is a negative regulator of katG expression
The linkage between furA and katG in all mycobacteria and changes in sensitivity to H2O2 in the M. smegmatis furA mutant are suggestive of a role for furA in katG expression. To determine whether furA regulates katG expression, we compared steady-state levels of KatG in the M. smegmatis furA::Kmr mutant JS106-1 and its furA+ parent mc2155. In the furA::Kmr background, KatG levels were markedly increased (Fig. 3A, lanes 1 and 2). A quantitative analysis of the steady-state KatG levels indicated a 4.2-fold increase in KatG in JS106-1 relative to mc2155 (Fig. 3A; P = 0.0001, anova). The overexpression of katG observed in JS106-1 was a direct result of the disruption of the furA gene, as plasmid pMsFurA (furA+) complemented the JS106-1 phenotype (Fig. 3A, lanes 2 and 3; P = 0.0001, anova). We attribute the reduction in KatG levels in JS106-1 [pMsFurA] beyond those observed in the furA+ wild-type strain mc2155 (Fig. 3A, lanes 1 and 3) to multicopy effects of plasmid-borne furA+. The less than wild-type levels of KatG in JS106-1 [pMsFurA] (Fig. 3A, lanes 1 and 3) were also in keeping with the hypersensitivity of the complemented strain to H2O2 (Table 1, rows A and C). Next, we investigated whether furA, in addition to KatG, affected steady-state levels of AhpC. When strains mc2155, JS106-1 and JS106-1 [pMsFurA] were tested, no significant differences in AhpC amounts were observed (Fig. 3B). These observations were also in keeping with the lack of detectable changes in susceptibility to CHP between mc2155, JS106-1 and JS106-1 [pMsFurA] (Table 1, rows A–C). In conclusion, the data presented indicate that FurA is a negative regulator of katG but not of ahpC in M. smegmatis.
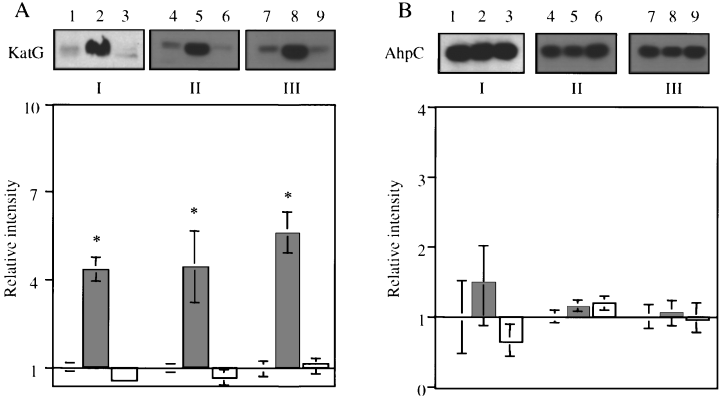
Steady-state KatG and AhpC levels in M. smegmatis strains grown in Sauton's minimal medium containing different concentrations of iron. Western blot analysis and band intensity quantification were performed using total protein extracts (5 µg) from wild-type mc2155 (furA+ lanes 1, 4 and 7), JS106-1 (furA::Kmr; lanes 2, 5 and 8), and JS106-1 [pMsFurA] (furA::Kmr[pfurA+]; lanes 3, 6 and 9) and antibodies that recognize mycobacterial KatG (A) and AhpC (B). Strains were grown in minimal medium supplemented with 0.5 µM Fe2+ (set I: lanes 1–3), 5 µM Fe2+ (set II: lanes 4–6) or 70 µM Fe2+ (set III: lanes 7–9). Western blots from representative gels are shown in each case. Graphs represent quantitative analyses of band intensities (see Experimental procedures) performed on independent triplicate cultures. Relative intensities of KatG and AhpC were normalized against wild-type mc2155 levels and expressed as mean ± SE values. Statistical analysis was performed on mean relative intensity under each condition tested. *, P < 0.005 (anova); mc2155 relative to JS106-1.
Analysis of the role of furA in katG and ahpC expression in response to oxidants and iron availability
We next investigated whether furA affected steady-state levels of KatG and AhpC upon exposure to oxidants. The absence of furA did not significantly affect KatG levels in cells treated with subinhibitory concentrations (125 µM) of H2O2 (Fig. 4A). This concentration of H2O2 was previously determined to provide optimal induction analysis (Dhandayuthapani et al., 1996). These results suggest that steady-state KatG levels do not change significantly in M. smegmatis upon H2O2 treatment at concentrations of this agent previously shown to induce ahpC and other polypeptides in this organism (Sherman et al., 1995; Dhandayuthapani et al., 1996). Furthermore, the derepression of katG in the furA::Kmr mutant was independent of peroxide stimulation (Fig. 4A). We also examined steady-state levels of AhpC after treatment with subinhibitory concentrations (125 µM) of the organic peroxide CHP. This concentration of CHP was previously determined to be optimal for induction studies (Dhandayuthapani et al., 1996). Treatment of mc2155, JS106-1 and JS106-1 [pMsFurA] with CHP resulted in an ≈ 1.5-fold increase in steady-state levels of AhpC over untreated wild-type levels (Fig. 4B; P = 0.0352, anova). However, as expected, this increase was independent of furA, as higher levels of AhpC were achieved in both furA+ and furA::Kmr mutant cells (Fig. 4B, lanes 2 and 5). These results are also consistent with the interpretation that furA does not affect steady-state levels of AhpC and is not involved in AhpC increases upon exposure to peroxides.

Steady-state KatG and AhpC levels in M. smegmatis strains grown in 7H9 and exposed to H2O2 and cumene hydroperoxide (CHP). Western blot analysis and band intensity quantification were performed using total protein extracts (25 µg) from wild-type mc2155 (furA+ lanes 1 and 4), JS106-1 (furA::Kmr; lanes 2 and 5) and JS106-1 [pMsFurA] (furA::Kmr[pfurA+]; lanes 3 and 6) using KatG (A) and AhpC (B) antibodies. Western blots from one representative gel are shown. Relative intensities of KatG and AhpC are mean ± SE values from at least three independent cultures. Statistical analysis was performed on mean relative intensity under each condition tested. *, P < 0.05 (anova); mc2155 relative to JS106-1.
As the mycobacterial FurA shows strong similarities to the iron uptake regulator Fur and other Fur-like elements (Deretic et al., 1997; Pagan-Ramos et al., 1998), we next tested whether furA regulated steady-state levels of KatG in response to iron. When mc2155, JS106-1 and JS106-1 [pMsFurA] were grown in conditions of low (0.5 µM), intermediate (5.0 µM) or high (70 µM) iron (Wong et al., 1999), no significant differences in KatG levels were observed in the context of furA repression of katG (Fig. 3A; set I, 0.5 µM Fe2+ set II, 5.0 µM Fe2+ set III, 70 µM Fe2+). In keeping with these observations, sensitivities to H2O2 of M. smegmatis mc2155 and its derivatives JS106-1 and JS106-1 [pMsFurA] remained unaffected when bacteria were grown in Sauton's minimal medium supplemented with various concentrations of iron (data not shown). Owing to similarities with the ferric uptake regulator Fur, we also examined whether inactivation of furA affected the production of siderophores in M. smegmatis, a known iron-regulated system (Dussurget et al., 1996). No differences in siderophore secretion between the furA+, furA::Kmr and an ideR::Kmr strain were observed on CAS medium supplemented with 0.5 µM Fe2+ (6 mm secretion zones; data not shown). However, on CAS medium supplemented with 70 µM Fe2+ (Fig. 5A), high iron repressed siderophore secretion in wild-type M. smegmatis mc2155 (0 mm secretion zone), but not in the ideR mutant (6 mm secretion zone). The siderophore repression by iron also remained unaltered in the furA::Kmr mutant strain JS106-1 (Fig. 5A). Thus, in contrast to IdeR, FurA most probably plays no major role in the aspect of iron regulation in M. smegmatis that involves siderophore secretion.
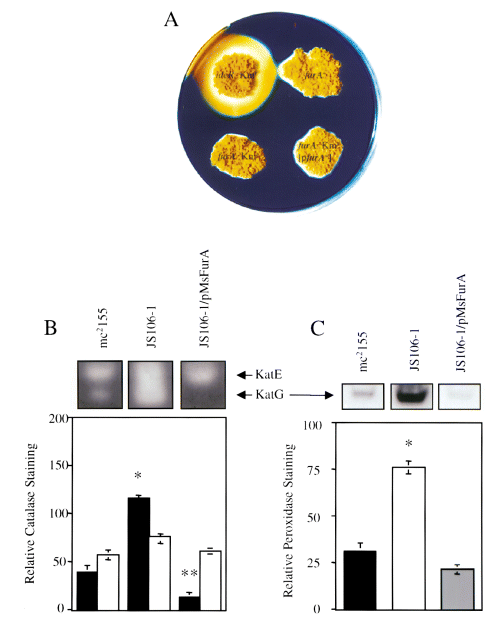
Comparison of siderophore secretion and peroxidase and catalase activity zymograms in furA+ and furA mutants from M. smegmatis.
A. Siderophore secretion from wild-type strain mc2155 (furA+), JS106-1 (furA::Kmr), JS106-1 [pMsFurA] (furA::Kmr[furA+]) and SM3 (ideR::Kmr) was analysed on CAS medium containing 70 µM Fe2+. Derepression of siderophore secretion produces a yellow halo. Secretion zones were measured from the colony edge.
B and C. Relative peroxidase and catalase activities of total protein extracts from mc2155, JS106-1 and JS106-1 [pMsFurA] after activity gel staining. Total protein extract (50 µg) was loaded and run on non-denaturing polyacrylamide gels and stained for catalase (B) or peroxidase (C) activities as described in Experimental procedures. In (B), KatE activity is the upper, slower migrating band (open bars), and KatG activity is the faster migrating, lower band (filled bars). Activity staining from one representative gel is shown. Relative staining is the mean ± SE from at least three independent cultures. Statistical analysis was performed on mean values from relative catalase or peroxidase activity staining between mc2155 and either JS106-1 or JS106-1 [pMsFurA]. *, P < 0.0001 (anova); mc2155 relative to JS106-1; **, P < 0.05 (anova); mc2155 relative to JS106-1 [pMsFurA].
Comparison of the effects of furA inactivation on KatG and KatE expression levels
M. smegmatis synthesizes two enzymes with catalase activity: KatG, which functions as a catalase and peroxidase (Zhang et al., 1992; Heym et al., 1993); and KatE, with catalase activity only (Bartholomew, 1968; Dussurget et al., 1996). Because inactivation of furA increased the expression of katG (3, 4) and altered the susceptibility of M. smegmatis to H2O2 (Table 1), we next examined whether furA also regulated the expression of the katE gene, encoding the heat-stable catalase. When protein extracts from wild-type mc2155, JS106-1 (furA::Kmr) and JS106-1 [pMsFurA] (furA::Kmr/furA+) were subjected to zymogram analysis for catalase activity, no significant differences were observed in katE-encoded catalase activity (Fig. 5B). This was in contrast to a significant increase in katG-encoded catalase activity observed in JS106-1 compared with wild-type mc2155 (Fig. 5B; P < 0.0001, anova). The increase in katG-encoded catalase activity stain was a direct result of furA disruption, as complementation of JS106-1 with pMsFurA reversed the relative increase in KatG catalase staining back below wild-type levels (Fig. 5B; P = 0.0175). These results are consistent with the relative increase and decrease in steady-state KatG levels observed by Western blot analysis in JS106-1 and JS106-1 [pMsFurA] strains respectively (3, 4). The increase in katG expression in JS106-1 also resulted in increased peroxidase activity staining compared with wild-type mc2155 (Fig. 5C; P < 0.0001). These results suggest that, unlike katG, katE is most probably not regulated by furA. Furthermore, the increase in katG expression after disruption of furA results in an increase in both catalase and peroxidase activities of KatG, but does not indirectly alter KatE catalase activity.
Sensitivity to INH and furA
Inactivation of furA decreases the susceptibility of M. smegmatis to H2O2 (Table 1) and increases steady-state levels of KatG (Figs 3A and 4A). As KatG has been postulated to activate the prodrug INH, transforming it intracellularly into an active form (Wengenack et al., 1998; Lei et al., 2000), we tested whether the loss of katG repression in the furA::Kmr mutant would also affect the susceptibility of M. smegmatis to INH when grown in 7H9 medium. When JS106-1 (furA::Kmr) was compared with mc2155 (furA+), a significant increase in susceptibility to INH was observed (Table 1, rows A and B; P = 0.0001, anova). Introduction of the complementing plasmid pMsFurA into JS106-1, which reduced katG expression to below wild-type levels (3, 4), partially reversed the INH sensitivity phenotype (Table 1, rows A–C; P = 0.0001). The inability to complement the INH phenotype fully (Table 1, rows A and C), in contrast to the full complementation of KatG levels (3, 4) and H2O2 sensitivity (Table 1), suggests that furA may affect or regulate additional genes that respond to multicopy complementation differently from katG. Alternatively, excessive repression of katG may render M. smegmatis increasingly susceptible to some aspects of INH antimycobacterial action that depend on the generation of reactive oxygen intermediates (Shoeb et al., 1985) normally detoxified by KatG. Consistent with a lack of effect of iron on KatG activity (Fig. 3A), no additional changes in INH susceptibility were observed in mc2155, JS106-1 and JS106-1 [pMsFurA] when bacteria were assayed under conditions of low, intermediate or high iron (data not shown).
Discussion
In this work, a regulatory role has been established for the mycobacterial furA gene. The data presented show that FurA acts as a negative regulator of katG in M. smegmatis. Although, at this stage, it cannot be stated definitively whether the regulation of katG by FurA is direct or indirect, the derepression of katG as a result of insertional inactivation of furA cannot be attributed to a polar effect on the downstream katG gene based on the following considerations. (i) If the insertion were polar, KatG levels would have been reduced and not increased in the mutant, contrary to our observations. (ii) More importantly, the chromosomal mutation in furA was complemented in trans and its phenotypic consequences reversed by the introduction into the furA::Kmr mutant of a plasmid-borne furA+. (iii) The genetic complementation also rules out the possibility of increased katG expression resulting from promoter activity from the Kmr cassette reading through the katG gene.
Based on the invariant furA–katG linkage in mycobacteria, as reported in M. tuberculosis, M. leprae and M. marinum (Deretic et al., 1997; Pagan-Ramos et al., 1998) and further extended in this work to include M. smegmatis, M. avium, M. aurum, M. fortuitum and M. xenopi, it is likely that the regulatory relationships between furA and katG are universal in mycobacteria. These relationships may potentially be extended to other actinomycetes, as a similar organization has been reported in Streptomyces reticuli (Zou et al., 1999) and Streptomyces coelicolor (Hahn et al., 2000). The regulation of catalase genes by fur homologues has also been reported even when the genes are not linked in a number of bacteria including Bacillus subtilis (Bsat et al., 1998), Pseudomonas aeruginosa (Hassett et al., 1997) and Campylobacter jejuni (van Vliet et al., 1998; 1999). Iron-related regulation of the catalase–peroxidase gene katG has been observed previously in M. smegmatis (Dussurget et al., 1996). However, inactivation of ideR affects katG in a different way from furA, as steady-state levels of KatG are reduced in ideR mutant cells (Dussurget et al., 1996), whereas furA inactivation increases KatG levels. The molecular mechanism responsible for katG regulation by IdeR is not known and could be indirect, potentially via changing intracellular iron concentrations or other downstream events caused by the elevated expression of siderophores in ideR mutant cells. Indirect evidence for the role of iron in the regulation of a closely related catalase–peroxidase gene by a fur homologue has been demonstrated in the case of furS regulation of cpeB in S. reticuli (Zou et al., 1999). However, these studies were carried out using plasmid constructs in lieu of a bona fide furS chromosomal mutant. In addition, iron-mediated (as well as Ni2+, Mn2+ and Zn2+) regulation of catC by furA has been demonstrated in the closely related species S. coelicolor (Hahn et al., 2000). Although we have not found evidence in support of a role for exogenous iron in the regulation of katG by furA, this does not preclude the possibility that intracellular iron or another redox active transition metal may affect katG expression. Precedents for such relationships have been observed in other organisms, including the manganese-dependent (in addition to iron) regulation of katA by PerR in B. subtilis (Bsat et al., 1998) and the zinc-dependent regulation exhibited by Zur in Escherichia coli (Patzer et al., 1998). Regardless of the specifics of the potential metal involved or other type of redox sensing by FurA, it appears that regulation of the oxidative stress response by Fur homologues is a widespread mechanism in mycobacteria and other organisms.
The biological properties of strains with inactive furA suggest several intriguing phenotypic consequences with broader implications for the physiology and pathogenic characteristics of mycobacteria. First, the furA::Kmr strain displays higher resistance to hydrogen peroxide, thus representing an unexpected improvement upon the wild-type strain in detoxification of H2O2. Secondly, another notable change is the increased susceptibility of the furA::Kmr strain to INH. Both these phenomena can be explained, at least in part, by the increased levels of KatG and the resulting increase in total catalase and peroxidase activities.
The finding that furA regulates the expression of katG in M. smegmatis potentially sheds light on some of the curious aspects of the evolution of the pathogenic mycobacteria. For example, M. leprae has an inactivated furA gene (Deretic et al., 1997; Pagan-Ramos et al., 1998), a phenomenon that, at least at some stage in the speciation of this pathogen, could have enhanced its ability to detoxify peroxides and perhaps survive in the host. Similar selective pressures might have existed in M. tuberculosis as elimination of functional oxyR took place. The effects of the deregulation of oxidative stress response in these organisms must have provided some selective advantage, as only the frank human pathogens M. leprae and M. tuberculosis have undergone such irreversible changes, in sharp contrast to other opportunistic or non-pathogenic mycobacteria.
Experimental procedures
Bacterial strains, media and growth conditions
JS106-1(furA::Kmr), VD1865-6 (ahpC::Kmr) (Zhang et al., 1996) and SM3 (ideR::Kmr) (Dussurget et al., 1996) are isogenic derivatives of M. smegmatis mc2155 (Snapper et al., 1990). All transformations performed in E. coli were in DH5α. M. smegmatis was grown in Middlebrook 7H9 broth or 7H10 agar (Difco) supplemented with ADC (10% bovine serum albumin fraction V, dextrose and sodium chloride), 0.2% glycerol and 0.05% Tween 80 or in a modified Sauton minimal medium (Dussurget et al., 1996). When required, 0.75% noble agar (Difco) was added to 7H9 or Sauton media for soft agar. For preparation of Sauton, glassware was soaked with 0.2 M HCl and media treated with 5.0 g l−1 Chelex 100 resin (Bio-Rad) to remove metal contaminants before the addition of trace elements and the iron source, ferrous ammonium citrate. E. coli was grown in Luria–Bertani (LB) medium (Difco). When required, kanamycin sulphate (Km; Sigma) and hygromycin B (Hyg; Boehringer Mannheim) were added at 25 µg ml−1 and 50 µg ml−1, respectively, for M. smegmatis or 50 µg ml−1 and 200 µg ml−1, respectively, for E. coli. For exposure of M. smegmatis to peroxides, strains were grown in 7H9 to an optical density at 600 nm (OD600) of 0.5, aliquoted into 5 ml cultures and exposed to 125 µM H2O2 or 125 µM CHP (Sigma) for 1 h in a 37°C shaker. Detection of siderophore secretion was performed on CAS medium (Dussurget et al., 1996).
Cloning and recombinant DNA techniques
Inverse PCR was used to clone the region upstream of katG in M. smegmatis based on the sequence of the 5′ end of M. smegmatis katG (Billman-Jacobe et al., 1996). M. smegmatis genomic DNA (2.5 µg) was digested to completion with SphI, purified by Qiaex II (Qiagen) and self-ligated in a 25 µl reaction mixture. Inverse PCR was carried out with 5 µl of the ligation reaction mixture and primers specific to the known portion of M. smegmatis katG, Msm-fur2 (5′-CCGGCGGGTTTCGGCCGCATC-3′), and a small 5′ region immediately upstream of katG, Msm-fur1 (5′-TTCCTTTCGGGAGTGGTGAAT-3′). A single band corresponding to an 800 bp PCR product was cloned into pCR2.1 (Invitrogen) and sequenced to obtain the complete nucleotide sequence of M. smegmatis furA. A SacI-linearized fragment containing furA::Kmr from pSM243/MsfurA::Kmr was used for the disruption of furA in M. smegmatis mc2155 by allelic exchange. pSM243/MsfurA::Kmr was constructed as follows: primers Ms-fur10A (5′-ACGAGCTCTGCAGAAGGATCCACTGAAATTCGATGC-3′; underlined sequence SacI site) and Ms-fur11 (5′-CTACTAGC TAGCCAGCAGACTAGTGTTGTCGCCGACGCGGATTCGTAGCG-3′; underlined sequence SpeI site), and primers Msfur-12 (5′-CTGCTGGCTAGCTAGTAGACTAGTGTGGTCTGCCGCGCGTGCGGCGACATC-3′; underlined sequence SpeI site) and Msfur-13A (5′-GGTGAGCTCCCACTCGTTGCCGTACAGGATCCCAG-3; underlined sequence SacI site) were used to PCR amplify fragments containing the upstream sequence and 5′ half, and 3′ half and downstream sequence, respectively, of M. smegmatis furA. These fragments were ligated together, digested with SacI and cloned into the SacI site of pSM243 (provided by Dr I. Smith). An NheI–SpeI fragment encoding the Kmr cassette from pMV206 (Stover et al., 1991) was subsequently cloned into the engineered SpeI site present in furA, resulting in pSM243/MsfurA::Kmr. DNA from pSM243/MsfurA::Kmr was linearized with SacI, gel purified to enrich for fragments containing M. smegmatis furA::Kmr and electroporated into M. smegmatis mc2155 as described previously (Jacobs et al., 1991). Transformants resulting from double cross-over to exchange furA+ for furA::Kmr in M. smegmatis mc2155 were selected on 7H10 medium containing Km and screened by PCR using primers Ms-fur1 and Ms-fur5 (5′-CCGAGGCCGTCGGAGGAA-3) that flank the Kmr cassette. Plasmid pMsFurA was used for complementation of furA::Kmr in M. smegmatis JS106-1 and was constructed as follows: a DNA fragment containing furA was PCR amplified from M. smegmatis mc2155 using primers Ms-fur5 and Ms-fur9 (5′-CTTCTGCAGGATCTTCAGATTGAGCTGATT-3′) and cloned into pCR2.1. A BamHI–PstI fragment containing furA was then ligated into the BamHI–PstI-digested E. coli–mycobacterium shuttle vector pOLYG (Hygr) to create pOLYG/MsfurA. Finally, the XbaI–NheI-digested xylE reporter gene from pHSX-1 (Curcic et al., 1994) was cloned into the XbaI polylinker site of pOLYG/MsfurA to create pMsFurA. JS106-1 transformants containing pMsFurA were selected on 7H10 medium containing Hyg and screened for the presence of xylE, detected as yellow colonies upon spraying with 100 mM catechol (Curcic et al., 1994). For the amplification of furA from other mycobacterial species, two degenerate primers were used: FurAZooL [5′-GTCGGCGACAACCACCACCAC(AG)T(CG)GT(CG)ACG-3′) and FurAZooR [5′-GCCA(CG)AGCAG(CG)CGGCG(CG)GCCTTCAGT(AG)CG-3′]; residues in parenthesis indicate degenerate positions.
DNA extraction and Southern analysis
Mycobacterial genomic DNA was prepared as described previously (Jacobs et al., 1991). For Southern analysis, 4 µg of genomic DNA was digested overnight with PstI (Gibco BRL), separated by electrophoresis on a 0.8% agarose gel, transferred onto a Duralon-UV membrane (Stratagene) and used in subsequent high-stringency hybridization and washing steps (Pagan-Ramos et al., 1998). A furA-specific probe from M. smegmatis was generated by random-primed labelling (Gibco BRL) with [α-32P]-dCTP (3000 Ci mmol−1; NEN Dupont) using PCR products generated with oligonucleotides Ms-fur1 and Ms-fur5.
Zone inhibition assays
A modified disc inhibition assay (Rosner, 1993) was used to determine the relative sensitivities of M. smegmatis derivatives to H2O2, CHP and INH. M. smegmatis strains were grown to mid-exponential phase (OD600 = 0.5) in 7H9 or Sauton media containing low (0.5 µM), moderate (5 µM) or high (70 µM) Fe2+. A sample of 0.2 ml of cells was added to 3 ml of tempered 7H9 or iron-supplemented Sauton soft agar, poured onto the respective agar plates and allowed to solidify. Sterile quarter-inch-diameter BBL blank paper disks (Fisher) were added to the centre of bacteria-overlaid plates and saturated with 10 µl of a 2% solution of H2O2 or CHP or a 1 mg ml−1 solution of INH. Plates were incubated for 3 days at 37°C before zones of growth inhibition were measured.
Immunoblot analysis
Crude cell extracts of strains grown to mid-exponential phase (OD600 = 0.5) were prepared by homogenization with a mini bead beater and 0.1 mm zirconia beads (Biospec Products) for 1 min. Cell debris and beads were removed by centrifugation, and supernatants were measured for total protein. Aliquots of 25 µg (from 7H9-grown bacteria) or 5 µg (from bacteria grown in Sauton's medium) of total protein from M. smegmatis derivatives were separated on SDS−11% polyacrylamide gels, transferred to Immobilon-P membranes (Millipore) by electroblotting and probed using rabbit antiserum to M. tuberculosis KatG (from Dr C. Barry) or AhpC (Pagan-Ramos et al., 1998). Goat anti-rabbit immunoglobulin G conjugated to horseradish peroxidase (Pierce) was used as the secondary antibody. Bound antibodies were visualized using ECL (NEN Research Products) as recommended. NIH image (version 1.62; National Institutes of Health) was used to quantify the relative intensity of KatG or AhpC proteins.
Enzyme activity stains
Catalase and peroxidase activities were assayed as described previously (Wayne and Diaz, 1986). Briefly, 50 µg of total unboiled protein extract were loaded on a 7.5% non-denaturing polyacrylamide gel and run at 100 V at 4°C. For catalase staining, polyacrylamide gels were soaked in 5 mM H2O2, briefly washed in water and incubated in a solution containing 2% ferric chloride and 2% potassium ferricyanide. Catalase activity was visible as a clear area on a green-stained background. For peroxidase staining, polyacrylamide gels were soaked in a solution of 0.5 mg ml−1 diaminobenzidine prepared in 5 mM H2O2. Peroxidase activity was visible as a brown band on an achromatic background. NIH image was used to quantify the relative intensity of catalase or peroxidase staining from polyacrylamide gels.
Statistical analysis
All statistical analyses (analysis of variance and Fisher's protected least significant difference) were performed with anova (version 1.11; Abicus Software).
Nucleotide sequence accession numbers
The sequences reported here have been deposited in GenBank with the following accession numbers: (i) AF012631for M. smegmatis furA; (ii) AF092559 for M. avium furA–katG partial sequence; (iii) AF092558 for M. aurum furA–katG partial sequence; and (iv) AF092560 for M. xenopi furA–katG partial sequence.
Acknowledgements
We thank C. Barry for KatG antibodies, I. Smith for strain SM3, and E. Pagan-Ramos for useful comments. This work was supported by National Research Service Awards AI10278 to T.C.Z and AI10022 to J.S., and by grant AI42999 from the National Institute of Allergy and Infectious Disease to V.D.