High genetic diversity and no inbreeding in the endangered copper redhorse, Moxostoma hubbsi (Catostomidae, Pisces): the positive sides of a long generation time
Abstract
The evolutionary potential of a species is determined by its genetic diversity. Thus, management plans should integrate genetic concerns into active conservation efforts. The copper redhorse (Moxostoma hubbsi) is an endangered species, with an endemic distribution limited to the Richelieu River and a short section of the St Lawrence River in Quebec, Canada. The population, gradually fragmented since 1849, is characterized by a decline in population size and a lack of recruitment. A total of 269 samples were collected between 1984 and 2004 and genotyped using 22 microsatellite loci, which indicated that these fish comprise a single population, with a global FST value of only 0.0038. Despite a small census size (∼500), a high degree of genetic diversity was observed compared to common values for freshwater fishes (average number of 12.5 alleles/locus and average HO of 0.77 ± 0.08). No difference was observed between expected and observed pairwise values of relatedness (rxy: −0.00013 ± 0.11737), suggesting an outbred population. Long-term Ne was estimated at 4476 whereas contemporary Ne values ranged from 107 to 568, suggesting a pronounced yet gradual demographic decline of the population, as no bottleneck could be detected for the recent past. By means of simulations, we estimated Ne would need to remain at more than ∼400 to retain 90% of the genetic diversity over 100 years. Overall, these observations corroborate other recent empirical studies confirming that long generation times may act as a buffering effect contributing to a reduction in the pace of genetic diversity erosion in threatened species.
Introduction
Active management is required to prevent the extinction of threatened species, including actions such as supportive breeding programmes (i.e. the propagation and release of offspring from a subset of wild breeders) or translocation of individuals from other populations. Since the evolutionary potential of a species is partly governed by its genetic diversity, management plans must be concerned with genetic objectives (Frankham et al. 2002). The primary challenge is to integrate genetic concerns into active conservation efforts and to consider the utility of the genetic information obtained as potentially useful for future conservation efforts (DeSalle & Amato 2004). Genetic considerations can greatly contribute to the success of such recovery plans in various ways (Hedrick & Kalinowski 2000; Tallmon et al. 2004a). Strong population structure would indicate the existence of different reproductive sites and nurseries, thus implying the necessity to protect those essential habitats. In such cases, individuals should not be translocated or used for breeding programs between genetically distinct locations in order to avoid decreases in local adaptation (Tallmon et al. 2004a). Loss of genetic diversity and an increase in inbreeding are two additional genetic issues to be considered. Ballou & Lacy (1995) noted that the avoidance of inbred crosses in a supportive breeding programme may reduce the rate of loss of genetic diversity by 20–30% in some cases. Moreover, both historical bottlenecks and current population size of threatened species have been found to explain observed contemporary levels of genetic diversity and heterozygosity (e.g. Westemeier et al. 1998; Hoelzel 1999; Hauser et al. 2002; Johnson et al. 2004; Knaepkens et al. 2004; Pastor et al. 2004). It is therefore imperative to detect natural population declines at the earliest stage possible, and to monitor the flux in the effective population size, and its effect on the population's overall genetic health.
During a period of population size reduction, the rate of loss of genetic diversity could be influenced by demographic stochasticity, variance in reproductive success and disrupted gene interactions (Hoelzel 1999). The impact of a population reduction will also depend on life history traits that affect growth rate (e.g. iteroparity, overlapping generations and lifespan of long-lived species), as well as the severity and pattern of the bottleneck experienced and population recovery (England et al. 2003; Weber et al. 2004). Of particular interest are recent empirical observations that all factors contributing to a reduction in genetic diversity may potentially be buffered by long generation times, as observed among different vertebrate species (Amos & Balmford 2001; Kuo & Janzen 2004; Goossens et al. 2005).
Natural populations of freshwater fishes are increasingly threatened by anthropogenic habitat destruction and fragmentation, contributing to a reduction of population sizes and increasing the risk of extinction due to the stochastic nature of the environment (Cambray & Bianco 1998; Ricciardi & Rasmussen 1999; Frankham et al. 2002). Dams, agriculture and urban development degrade water quality, encourage the introduction of exotic species by modifying the environment, and increase habitat fragmentation. These represent the main current threats to the long-term survival of freshwater fishes (Cambray 2000; Cooke et al. 2005). Members of the catostomid family are distributed over North America and Asia, and inhabit a wide range of habitats. Many members of this family, such as the razorback sucker (Xyrauchen texanus) and the robust redhorse (Moxostoma robustum), are threatened by common factors: migration barriers and habitat degradation by urbanization and agriculture (Cooke et al. 2005). Unfortunately, little conservation effort has been implemented for the conservation of these less economically important, ‘nonrecreational’ freshwater fishes (Cooke et al. 2005).
The copper redhorse (Moxostoma hubbsi) is endemic to Canada, and its current geographic distribution is limited to the St Lawrence and Richelieu rivers in Quebec (Fig. 1). The species is characterized by a long lifespan (30 years) and late maturation age (minimal age at maturity being 10 years), as well as a very specialized diet, which is almost entirely composed of mollusks (Mongeau et al. 1992). Only two breeding sites are known for the whole species, both located in the Richelieu River (Saint-Ours and Chambly), and these have been progressively isolated by a dam since 1849. The original cribwork structure was frequently damaged and likely not fully impassable; it was rebuilt in 1969 and equipped with radial gates. Fish passage is possible for a few weeks during high spring discharge, typically in early spring, before the copper redhorse spawning migration period in June (Dumont et al. 1997). Historically, the copper redhorse was much more abundant relative to other catostomids (Courtemanche & Elliot 1985). However, the census size for the whole species was estimated to be fewer than 500 adult individuals in 2000, based on mark-recapture studies in feeding habitats (Vachon & Chagnon 2004). Moreover, the species is characterized by an extremely reduced reproductive success rate, translating into a lack of recruitment (Vachon & Chagnon 2004); only one young of the year has been captured each year in the Richelieu River since 1998 (Vachon 2002; COSEWIC in press). This lack of recruitment can hypothetically be explained by the late spawning time (which coincides with a peak in fertilizer contamination) of the copper redhorse compared to other catostomids inhabiting the Richelieu River (Mongeau et al. 1986). Consequently, the copper redhorse was listed in 1996 as ‘Vulnerable’ in the IUNC Red List of Threatened Species, in 1999 as ‘threatened’ under the Act respecting threatened and vulnerable species in Québec, and in 2004 as ‘threatened’ by the COSEWIC (Committee on the Status of Endangered Wildlife in Canada).
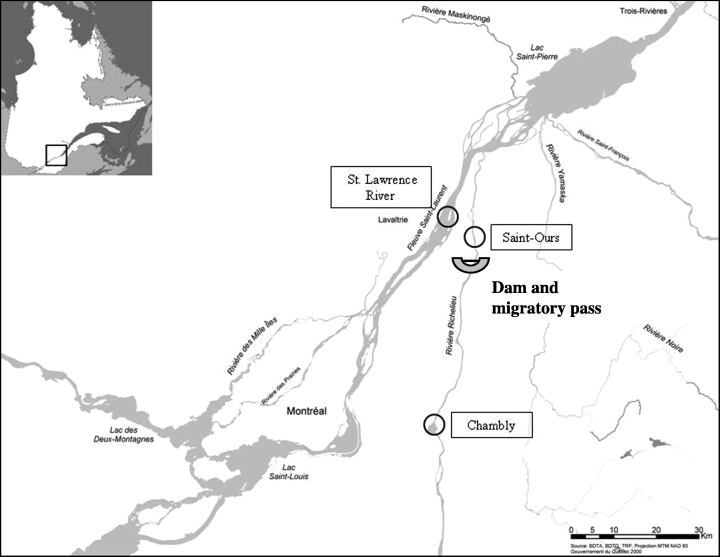
Geographic distribution of the copper redhorse in Quebec, Canada. Black circles represent the sampling sites (Saint-Ours, Chambly and St Lawrence River).
A recovery plan, involving numerous governmental and nongovernmental agencies (see Comité de rétablissement du Chevalier Cuivré 2004), was undertaken in 1995 with the ultimate goal of avoiding the extinction of the copper redhorse. These efforts have led to an increase in the knowledge of the biology of the species, as well as to the development of artificial rearing procedures (Branchaud et al. 1993, 1995; Branchaud & Gendron 1993), the creation of a sanctuary (Pierre-Étienne-Fortin sanctuary, 1999) to protect the main spawning site in the Richelieu River, Chambly (Fig. 1), and the construction of a migratory pass in 2001 at the Saint-Ours dam, re-establishing the connection between the two sections of the Richelieu River. Finally, a supportive breeding programme was initiated in 2004, and is expected to continue for the next 10 years (Bernatchez 2004).
Within the context of this recovery plan, the main objectives of this study were to (i) test the hypothesis that, given its highly reduced census size, the copper redhorse is characterized by low genetic diversity, high levels of inbreeding and small effective population size; (ii) test the null hypothesis of no genetic differentiation among samples of copper redhorse collected throughout the current distribution range of the species; and (iii) explore the future evolution of genetic diversity under different demographic scenarios by means of simulations.
Materials and methods
Sample collection
Noninvasive samples (preserved bones, scales, and pelvic fin clips preserved in 95% ethanol) were collected at three locations: Chambly and Saint-Ours, which are the two known spawning sites in the Richelieu River, and the Lavaltrie-Contrecoeur section of the St Lawrence River, where no copper redhorse were captured between 1973 and 1998, despite exhaustive fishing efforts in the area (Fig. 1). Samples were obtained between 1984 and 2004 from various sources, including commercial fishery accidental by-catch in the St Lawrence River. In total, 269 fish were available for analysis (Table 1). Fish length varied between 480 and 780 mm, representative of the size range for adult individuals (Vachon & Chagnon 2004). The sex ratio F/M observed for the St Lawrence group was globally 1.3 (Vachon & Chagnon 2004), indicating a slightly more abundant presence of females, even though a sex ratio in favour of the males is generally observed at reproduction sites, possibly because the males migrate before the females (Branchaud et al. 1993, 1995) or are more active (Pierre Dumont, biologist with the Société de la faune et des parcs du Québec, personal communication).
DNA extraction and genotyping
Genomic DNA extraction for fin tissue samples was performed using DNeasy Tissue Kit (QIAGEN), following the standard procedure for animal tissue samples provided by the manufacturer. DNA from scale samples was extracted using the method described in Nielsen et al. (1997), with the exception that scale digestion occurred for 2 h at 37 °C, and DNA was concentrated to a final volume of 50 µL using Microcon-30 microconcentrators (Amicon). DNA from bone samples was extracted as detailed in Hagelberg (1994), with the exception that bones were ground using a stone grinder, of which only 0.1 g was added to 2 mL of digestion buffer [0.47 m EDTA pH 8.0, 0.5% of SDS, 0.04 mg of proteinase K], and incubated overnight at 56 °C with constant agitation. Total digestion extracts were used for phenol–chloroform DNA extraction, as in Olsen et al. (1996), and concentrated twice using Microcon-30 microconcentrator (Amicon), diluted to a final volume of 50 µL and purified with QIAquick PCR Purification Kit (QIAGEN) to eliminate PCR inhibitors. Extractions for scale and bone samples were performed at different time periods using different laboratory benches period than those from fresh tissues, in order to minimize the risk of contamination.
Twenty-two loci were used for genetic analyses (see Table 2; Tranah et al. 2001; Lippéet al. 2004). PCR conditions and amplification profiles are as described in Lippéet al. (2004). For scale and bone samples, 45 amplification cycles were carried out in a 25-µL reaction, modified from Herrmann & Hummel (1994) and containing 2 µL (50–100 ng) of DNA extraction product, 2.5 µL of reaction buffer (10 mm Tris-HCl pH 8.3, 50 mm KCl, 2.5 mm MgCl2, 1% gelatin), 4 µg of bovine serum albumin (BSA), 57 µm of each dNTPs (except for dTTP; concentration 26.5 µm), 0.1 pmol of each primer, 0.5 U of Taq polymerase and 4.92 µm of Tamra-labelled dUTP. PCR products were separated on an 8% denaturing polyacrylamide gel and visualized on an FMBio II scanner (Hitachi), with the GeneScan®-500 [ROX]™ size standard (Applied Biosystems) for standardized size scoring. Three controls were performed to test for contamination: a blank control extraction in each set to verify the purity of the DNA extractions and the cleanliness of reagents and containers, an empty well in each PCR plate to check the purity of the PCR reagents, and an empty loading lane in each gel to look for carry-over contamination. A mixture of samples from various groups was also analysed together, to ensure concordance of results. No false amplifications were detected.
| Locus | N | A | H O | H E | F IS |
|---|---|---|---|---|---|
| ** Dlu4296 | 253 | 12 | 0.771 | 0.787 | 0.020 |
| ** Dlu405 | 236 | 9 | 0.682 | 0.775 | 0.118 |
| Mohu-Lav194 | 260 | 11 | 0.838 | 0.837 | −0.002 |
| Mohu-Lav200 | 264 | 17 | 0.803 | 0.823 | 0.023 |
| Mohu-Lav203 | 263 | 12 | 0.787 | 0.798 | 0.013 |
| Mohu-Lav211 | 261 | 13 | 0.828 | 0.855 | 0.032 |
| Mohu-Lav212 | 204 | 13 | 0.843 | 0.855 | 0.012 |
| Mohu-Lav213 | 200 | 9 | 0.785 | 0.761 | −0.033 |
| Mohu-Lav229 | 262 | 21 | 0.779 | 0.772 | −0.009 |
| Mohu-Lav237 | 259 | 16 | 0.876 | 0.900 | 0.026 |
| Mohu-Lav268 | 254 | 18 | 0.752 | 0.757 | 0.006 |
| Mohu-Lav270 | 260 | 14 | 0.777 | 0.881 | 0.117 |
| Mohu-Lav277 | 263 | 9 | 0.768 | 0.789 | 0.026 |
| Mohu-Lav286 | 259 | 9 | 0.633 | 0.631 | −0.005 |
| Mohu-Lav294 | 262 | 22 | 0.927 | 0.929 | 0.000 |
| Mohu-Lav296 | 264 | 23 | 0.920 | 0.934 | 0.014 |
| Mohu-Lav305 | 261 | 4 | 0.608 | 0.630 | 0.035 |
| Mohu-Lav306 | 200 | 7 | 0.780 | 0.809 | 0.034 |
| Mohu-Lav321 | 261 | 10 | 0.674 | 0.716 | 0.058 |
| Mohu-Lav329 | 263 | 9 | 0.692 | 0.751 | 0.078 |
| Mohu-Lav336 | 262 | 11 | 0.706 | 0.719 | 0.017 |
| Mohu-Lav347 | 263 | 6 | 0.734 | 0.720 | −0.020 |
- All primers are from Lippéet al. (2004), except **from Tranah et al. (2001).
Data analysis
Genetic diversity, Hardy–Weinberg and linkage disequilibrium tests. Standard genetic statistics were used to measure genetic variability; number of alleles (A), observed heterozygosity (HO), expected heterozygosity (HE) under Hardy–Weinberg assumptions, and FIS estimates were computed with msa (Dieringer & Schlötterer 2003). Deviations from Hardy–Weinberg equilibrium were examined at each microsatellite locus within each sample group using genepop version 3.4 (Guo & Thompson 1992; Raymond & Rousset 1995). Fisher exact test of linkage disequilibrium was performed between all pairs of loci using genepop, with 10 000 dememorizations, 10 000 batches and 10 000 iterations per batch. Critical significance levels were adjusted for multiple comparisons by using the sequential Bonferroni correction (α = p/n; Rice 1989).
Population structure. Nineteen loci were used for these analyses owing to the nonamplification of three loci from bone and scale extracted DNA (Mohu-Lav212, Mohu-Lav213, Mohu-Lav306). genepop was used to conduct exact tests of genic differentiation between groups at each locus using allelic distribution (Guo & Thompson 1992). Global and pairwise genetic divergence in allelic identity was quantified by estimating FST values using genetix 4.03 (Weir & Cockerham 1984; Belkhir et al. 1996–2004). Finally, a model-based Bayesian clustering method (structure version 2.1) was used to corroborate F-statistic results with regard to the number of estimated populations of copper redhorse present in the system (Pritchard et al. 2000). Three simulations were conducted for each model (k = 1–5) assuming admixture and correlated allele frequencies between populations with a 100 000 replications burn-in period and 1 000 000 MCMC replicates.
Relatedness. Values of relatedness between all pairs of individuals without regard to sampling groups were calculated using the estimator rxy using kinship 1.3.1 (Queller & Goodnight 1989). Expected unrelated and full-sib pairwise relatedness was simulated from observed allelic frequency distributions at 19 loci (10 000 simulated pairs of full-sibs using kinship), and the distribution of simulated full-sib relatedness was used to detect full-sibs within the natural population (P > 0.01; Blouin et al. 1996). Then, the simulated relatedness values were used to plot a graph showing the distribution of expected values under random mating, which was then compared to the observed distribution. A deviation from random expectation due to an excess of high rxy values could result from nonrandom mating among related individuals, and/or excess in the representation from a few families. In both cases, this would be suggestive of an excess of inbred individuals in the population.
Effective population size. We first estimated the long-term species effective population size (Ne) using the heterozygosity-based method (Ohta & Kimura 1973), which predicts that at mutation–drift equilibrium under the stepwise-mutation model (SMM), Ne should equal [(1/1 –HE)2 − 1]/8µ, where µ is the mutation rate and HE is the mean expected heterozygosity across all loci. As this method depends on equilibrium processes, which makes it heavily sensitive to mutation modeling, we also calculated the long-term Ne under the infinite allele model (IAM) according to the equation: Ne = H/4µ(1 – H). We calculated long-term Ne for each locus individually as well as for the mean expected heterozygosity across all loci, and used two different mutation rates; µ = 5 × 10−4, which is the rate most commonly applied in fishes, and 10−4, an average microsatellite mutation rate (Jarne & Lagoda 1996; Estoup & Angers 1998). Indeed, to our knowledge, mutation rates in the order of either 10−3 or 10−5 have not been reported nor used in fishes. Several estimates of contemporary Ne were calculated for comparative purposes (Waples et al. 1993). The linkage disequilibrium method produces a contemporary (parental generation) inbreeding effective population size estimate (Neb, effective number of breeding adults), generated from a single temporal sample. We used the method developed by Hill (1981) and Bartley et al. (1992), which is based on the expectation that smaller populations will accumulate more disequilibrium over time. It was performed on all pooled samples captured since 1999 (N = 165), with 22 loci, using the program NeEstimator version 1.3 (Peel et al. 2004), which measures disequilibrium (D*) with Burrow's composite measure of disequilibrium (Campton 1987). The linkage disequilibrium method is considered the more accurate estimate because (i) the methodology requirements are appropriate for the sampling employed in this study (> 90 individuals, > 6 loci: Waples 1991; Bartley et al. 1992); (ii) it is ideally used for small natural fish populations (Waples & Teel 1990); (iii) it avoids the underestimation of Ne associated with temporal methods when only one generation separates samples (Waples 1989; Tallmon et al. 2004b); and (iv) it does not appear to be seriously affected by a reduction in population size (Waples 2005). For comparison and exploration purposes, temporal methods were also applied to estimate variance Ne for the interval between historical and current samples (harmonic mean of the effective sizes in generations 0 through t − 1), by examining genetic drift and change in allelic frequencies over time. According to plan I (Nei & Tajima 1981), historical samples from Saint-Ours 1985 and Chambly 1984 were assigned as generation zero (0), while generation one (t) was assigned to all samples caught since 1999, utilizing 19 loci. With the generation time approximated at ∼10–15 years, and a lifespan of ∼30 years, the sample interval of 15 years represents roughly one generation. The first temporal method implemented in mcleeps (version 1.1) estimated Ne using Monte Carlo Likelihood, and 100 000 Monte Carlo replicates were calculated for each value of Ne (Anderson et al. 2000). The maximum value for Ne was set to 600, after testing different maximum values (200, 600, 1000) to make sure that the result was not sensitive to this parameter, so that the estimate was reliable. mlne software was used for the second temporal method estimating the maximum ‘pseudo-likelihood’, still using 600 as the maximum value for Ne after testing the sensitivity of the method, as well as for a third temporal method, the moment estimator (Nei & Tajima 1981; Wang 2001).
Bottleneck tests. We tested for recent and pronounced reduction in population size using two different methods. bottleneck 1.2.02 (Piry et al. 1999), which is purported to be particularly sensitive to recent population bottlenecks (0.8–4.0 Ne generations), examines summary statistics through comparisons of a population's heterozygosity excess (HE) to that which is presumed to be found at mutation–drift equilibrium (HEq). In such cases, allelic diversity is predicted to be lost faster than heterozygosity (Cornuet & Luikart 1996). As microsatellites apparently evolve under a model more similar to the SMM than the IAM, the SMM and a two-phase mutation model were used (TPM; Di Rienzo et al. 1994), with 95% SMM and 5% IAM, with 12% variance of multiple-step mutations (Luikart & Cornuet 1998; Estoup & Cornuet 1999; Spencer et al. 2000). A total of 5000 simulation iterations were conducted, as suggested by Piry et al. (1999). Bottleneck significance was tested with the standardized differences test (Cornuet & Luikart 1996). We also performed the ‘M’ test of Garza & Williamson (2001), which uses gaps in allelic frequency distribution to detect population bottlenecks whereby the number of alleles (k) experiences a faster reduction than the allele size range (r). Therefore, the ratio M (k/r) under the SMM should be smaller in bottlenecked populations (Garza & Williamson 2001). Parameter values were set to the following: ps (proportion of one-step mutations) = 90% and Δg (average size of non one-step mutations) = 3.5. We estimated the θ parameter (4Neµ = θ) using Ne calculated from the linkage disequilibrium method, and the mutation rate of 5 × 10−4 mutation/locus/generation (Estoup & Angers 1998). The critical ratio (MC) was calculated with critical_m software to test significance, such that 95% of the 10 000 simulations of an equilibrium population had M > MC.
Simulations. The conservation goal of maintaining 90% of the initial genetic diversity over a 100-year period (Frankham et al. 2002) is arguably a more realistic objective than the optimal Ne of at least 500, historically recommended by Franklin & Frankham (1998) to maintain genetic diversity and avoid inbreeding depression. bottlesim (version 2.6) was used to explore the evolution of genetic diversity in long-lived species with overlapping-generations for a 200-year period, and estimate the sustainable population size needed in order to meet this specific conservation objective (Kuo & Janzen 2003). Estimations were performed with different Ne values temporally stable over a period of 200 years, with other simulation parameters kept constant (lifespan = 30 years, age at maturity = 10 years, completely overlapping generations, dioecious reproduction, random mating, sex ratio 1 : 1, 200 years simulated, 1000 iterations).
Results
Intrasample genetic diversity
Levels of genetic diversity observed in the copper redhorse were unexpectedly high, given the low estimate of census size for the species (Table 2). All loci were moderate to highly polymorphic, with number of alleles per locus ranging from 4 to 23 (mean = 12.5), and observed heterozygosity across all samples ranging from 0.61 to 0.93 (mean = 0.77 ± 0.08). The null hypothesis of Hardy–Weinberg equilibrium was not rejected after correcting the significance level for the number of pairwise comparisons of populations and loci (n = 88; α = 0.001), with only three sample–locus combinations (DLU405 and Mohu-Lav270 in the St Lawrence River and Mohu-Lav296 in Saint-Ours) showing significant heterozygote deficiencies. Fisher's exact test for linkage disequilibrium between all pairs of loci revealed only one pair of loci showing significant linkage (DLU405 and Mohu-Lav237) after sequential Bonferroni correction (n = 231, α = 0.0002), and 30 if considering the standard level of significance of 5%.
Population structure
Using a significance threshold of 0.05, only 19 of 114 genic differentiation tests were significant. After sequential Bonferroni correction, significant heterogeneity of allelic distribution among samples was found in four comparisons only (n = 114; α = 0.0004): St Lawrence vs. Chambly (Mohu-Lav203; P = 0.00007), Chambly vs. Saint-Ours (1990–2004) (Mohu-Lav203; P = 0.00020 and Mohu-Lav237; P = 0.00029) and Chambly vs. Saint-Ours 1985 (Mohu-Lav347; P = 0.00027). A global FST value of only 0.0038 (P = 0.0001) was observed, which reflected a very weak level of differentiation among all pairwise comparisons (ranging between 0.0019 and 0.0073) (Table 3). The null hypothesis of no population structure throughout the whole species distribution range could therefore not be firmly rejected. This was corroborated by the results of structure, which revealed no detectable population structure among sample groups. Constant Bayesian posterior probabilities for only the single population model (k = 1), as well as a lack of stability of the alpha parameter even after the important burn-in period and run length, were all indicative of the absence of population structure (Pritchard et al. 2000). Therefore, the whole species is apparently represented by a single reproductive population.
| 1999–2004 | 1984 | 1985 | 1990–2004 | |
|---|---|---|---|---|
| St Lawrence (1999–2004) | 0 | 0.0044 | 0.0024 | 0.0019 |
| Chambly (1984) | 1 | 0.0072 | 0.0073 | |
| Saint-Ours (1985) | 0 | 1 | 0.0070 | |
| Saint-Ours (1990–2004) | 0 | 2 | 0 |
Relatedness
The distribution of pairwise values of relatedness provided further evidence (in addition to high levels of heterozygosity) that the copper redhorse population is outbred. The empirical distribution closely matched that expected distribution under random mating (mean of 0.00175 ± 0.10971), with a mean value of −0.00013 ± 0.11737 (range: −0.93258–0.97420), which is typical of an outbred population. Moreover, very little overlap was observed with the simulated distribution of full-sibs, suggesting the absence of an over representation of a few families in the population (Fig. 2). Using the simulated estimates of full-sib pairwise relatedness, the full-sib detection threshold (0.01) was determined to be 0.23, with only 2.85% of all pairs of samples individuals potentially representing full-sibs under this criterion.
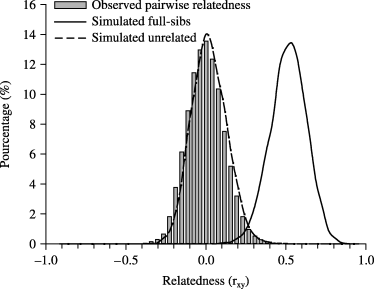
Percentage of pairwise relatedness (rxy; Queller & Goodnight 1989) values between all individuals sampled (histogram) compared to relatedness of simulated unrelated individuals (dashed curve) and full-sibs (solid curve).
Estimation of effective population size
Using a mutation rate of 5 × 10−4, the long-term species estimate of effective population size under the SMM(mean of 4476, with values ranging from 1577 to 57550 when using individual locus) was approximately one order of magnitude larger than the values obtained from the different methods used to obtain contemporary Ne estimates (Table 4). Under the IAM, long-term Ne was estimated to 1674 (individual locus estimates ranging from 852 to 7103), which is still higher than the current Ne estimates. By changing the mutation rate to 10−4, under both the SMM and IAM, the estimate of long-term Ne was even higher and increased to 22 380 and 8370, respectively. Among the contemporary Ne estimates, the linkage disequilibrium estimate was larger than the estimates from temporal methods. Yet, contemporary estimates were not strikingly different from census size estimates (N), resulting in unusually high Ne/N ratios for a fish species. Census size was estimated to ∼500, with the subsequent calculation of the Ne/N ratio ranging from 0.85 to 1.94 (when using the linkage disequilibrium method estimate; Ne = 480).
| Method | Source | Type and time period of estimate | Ne estimate |
|---|---|---|---|
| Heterozygosity-based | Ohta & Kimura (1973) | Long-term | 4476 (SMM); 1674 (IAM) |
| Linkage disequilibrium | Hill (1981) | Parental generation [General(t – 1) ∼ 1985–1990] | 480 [414–568] |
| Temporal methods | 1 generation | ||
| MCLEEPS | Anderson et al. (2000) | Harmonic mean of the | 260 |
| MLNE | Wang (2001) | effective sizes | 169 [107–357] |
| Moment-based | Waples (1989) | [General(0) to General(t– 1) ∼ 1985/1985–1990] | 116 |
Bottleneck tests
No evidence of population bottleneck was detected. According to bottleneck, no significant departure from mutation–drift equilibrium was observed for the entire population, based on the standardized differences test, under the assumption that all loci fit the TPM (T2 = −3.473; P = 0.00026) or the SMM (T2 = −6.241; P = 0.00000). Slight heterozygosity excess was observed for 11 out of 22 loci, which is expected to occur by chance in populations under mutation–drift equilibrium. Moreover, no evidence for population bottleneck could be inferred from the M ratio test either, with the average ratio among 22 loci (0.87) above the calculated critical ratio (MC) of 0.82 (only 6 loci under the threshold).
Simulations
Simulated levels of genetic diversity [observed number of alleles (OA) and observed heterozygosity (HO)] that would be retained after a 200-year period, with a stable population size characterized by an initial genetic diversity equal to the level we observed in empirical data, revealed that allelic richness declined much faster than HO (Fig. 3). If the copper redhorse population was to remain constant for the next 100 years with an effective population size of 480 (estimated from the LD method), 93.9% of the alleles present and 99.4% of the observed heterozygosity would be retained. To conform to general conservation goals of retaining 90% of the genetic diversity over a 100-year period, the effective population size would therefore need to remain at a value above ∼400, in which case 92.7% of the genetic diversity would be preserved.
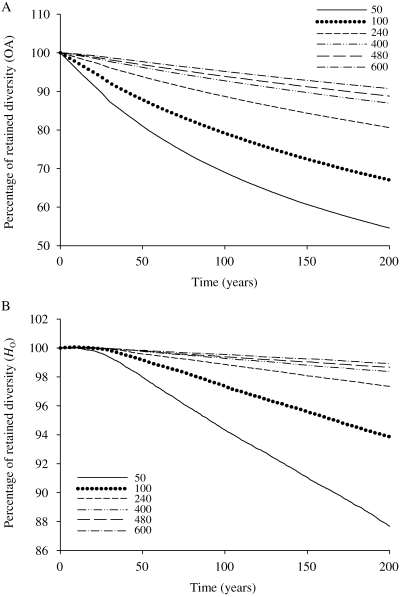
Evolution of genetic diversity over a 200-year period according to bottlesim simulations, assuming different Ne values kept constant over time with the same initial genetic diversity. (A) OA, observed number of alleles, (B) HO, observed heterozygosity. According to the percentage of retained observed number of alleles after 100 years, the minimal effective population size that should be aimed for the copper redhorse is around 400.
Discussion
The main objectives of this study were to (i) test the hypothesis that a highly reduced census size in the copper redhorse is associated with impoverished genetic diversity, high levels of inbreeding and small effective population size; (ii) test the null hypothesis of no genetic differentiation among samples of copper redhorse collected throughout the current distribution range of the species; and (iii) explore the future evolution of genetic diversity under different demographic scenarios by means of simulations. Despite a small census size of the entire adult population (∼500), the species exhibited a high degree of genetic diversity, well above average values commonly reported for freshwater fishes. No evidence of inbreeding was found, with a mean pairwise relatedness value which was typical for an outbred population. Yet, contemporary Ne estimates were roughly one order of magnitude less than the long-term Ne estimated. This, along with no evidence for a recent population bottleneck, suggested that the demographic decline of the copper redhorse occurred gradually, or at least not steeply enough to be detected by current analytical methods. By means of simulations, we estimated that in order to reach the general conservation goal to retain 90% of the genetic diversity over a 100-year period, the effective population size of the copper redhorse would need to remain at over ∼400, in which case 92.7% of the genetic diversity would be preserved. Overall, these observations corroborate other recent empirical studies suggesting that a long generation time may act as a buffer, which may contribute importantly to reducing the pace of genetic diversity erosion in threatened species.
Absence of population structure and high genetic diversity
No evidence of population structure was detected, indicating that the copper redhorse is composed of a single population. This discovery, associated with the only two known spawning grounds in the Richelieu River, could increase the species’ vulnerability to extinction in the event of any potential natural catastrophe or abrupt environmental changes. Yet, despite reduced access to one of the spawning grounds due to a dam construction, as well as population size reduction in the recent past, the copper redhorse was characterized by an unexpectedly high genetic diversity, showing all signs of an outbred population. Indeed, the average heterozygosity value observed in this study (0.77 ± 0.08) was substantially higher than most freshwater fishes [mean = 0.54 (± 0.25); DeWoody & Avise 2000].
The retention of such a high level of genetic diversity despite a currently small census size can be best explained by the long generation time of the copper redhorse in combination with the fact that demographic decline was apparently gradual. Given the actual effective population size estimate (ranging between 107 and 568), and the long-term effective size estimate under SMM with a mutation rate of 5 × 10−4 (4476), one explanation resides in the possibility that the gradual decrease of the long-lived copper redhorse population has allowed for a better retention of genetic diversity. Namely, a long lifespan of approximately 30 years and minimal age at maturity of about 10 years result in a long generation time that could buffer against the rate of loss of genetic variability. In addition, it has been reported for other long-lived species, such as the ornate box turtle (Terrapene ornate; Kuo & Janzen 2004) and the orang-utan (Pongo pygmaeus; Goossens et al. 2005), that the level of diversity observed may be influenced not only by long lifespan, but also by the pattern of demographic fluctuation (e.g. steep vs. gradual decline) experienced by populations. For example, high genetic diversity in the highly endangered razorback sucker (Xyrauchen texanus) was also observed and was explained by a gradual reduction in population size combined with life-history traits that are characteristic of long-lived species, as for the copper redhorse (Dowling et al. 1996a, b; Minckley et al. 2003; Dowling et al. 2005). Moreover, it is also possible that the recent population perturbations have not yet affected the species genetic diversity. All sampled individuals were mature adults, thus representing progeny resulting from reproductive events of the former generation that occurred 20–30 years ago. Despite the fact that the population has suffered an important demographic decline in recent years, old individuals that were analysed in this study apparently still represent most of the genetic diversity (both in terms of heterozygosity and occurrence of rare alleles) that characterized the species prior to demographic reduction.
High genetic diversity is typically suggestive of a genetically healthy population, and therefore represents a positive asset for the recovery of the species. On the other hand, extreme caution should be used when interpreting results like these, as they do not necessarily mean that the copper redhorse is unaffected by the reduction of Ne, or that it would remain unaffected in the future. Indeed, long lifespan and gradual demographic decline may mask an accelerated rate of genetic drift found in small populations (Kuo & Janzen 2004), so monitoring is essential to keep track of the evolution of genetic diversity in long-lived species such as the copper redhorse, which is experiencing a lack in recruitment and likely face the oncoming diminution of genetic diversity. Misinterpretation leading to the conclusions that the copper redhorse population is healthy and does not need further conservation efforts would in fact be misleading.
Effective population size estimate and Ne/N ratio
Long-term effective population size was calculated with different parameters (mutation models and rates) to provide stronger support for our conclusions. Temporal estimates of contemporary Ne were lower than those obtained from the linkage disequilibrium method. This could be due to the fact that, when the sampling scheme is according to plan I, variance effective population size estimated by temporal methods examines the variance of allele frequency between generations [General(0) to General(t − 1)], while the linkage disequilibrium estimates the inbreeding effective size of the parental generation [General(t – 1)] (Waples 2005). These two estimates might therefore be different if the size of the population is not constant. Given the recent demographic history of the copper redhorse, it is therefore understandable that the estimate from linkage disequilibrium method was higher than temporal estimates. Neither the consequences nor the signals of inbreeding due to shrinking population size may appear for several generations following a decline, resulting in a false sense of security when considering large estimates of inbreeding effective population size (Crow & Denniston 1988; Crandall et al. 1999; Schwartz et al. 1999). Moreover, large inbreeding effective population size, low census size and smaller variance effective population size, as we observed for the copper redhorse, may be indicative of a historically large population with recent population reduction (Templeton & Read 1994; Gerber & Templeton 1996). We must also consider that having only one generation separating our temporal samples implies that the harmonic mean of the effective sizes between generation 0 through t– 1 represents an extremely short time period (∼1985–90), where not much changes in allele frequency could possibly be observed relative to sampling errors, such that temporal methods are not extremely reliable for this case. Although the Ne estimates varied considerably between methods, their main outcome was similar: contemporary Ne is comparable to current census size but at least one order of magnitude lower than the long-term Ne estimate.
The estimated Ne/N ratio was close to one, which was surprising given that Frankham (1995) had found a mean ratio of 0.11 across species, and could be seen as an additional indicator of a genetically healthy population (Mace & Lande 1991; Nunney & Campbell 1993; Frankham 1995). Again, this result can be explained by the period of time that each estimate applies (Waples, 2005). As explained above, the contemporary Ne estimate given by the linkage disequilibrium method is not statistically representative of the current generation, but the parental generation that created our sample, that is adult individuals that were living 20–30 years ago when the census size was likely substantially higher than today. The census size estimated in 2000 represents the actual number of adults in the population. Assuming that the population of copper redhorse has been through a reduction in population in size during the preceding years, the Ne estimate from the linkage disequilibrium method would be higher than what would be found in the current generation, thus creating a disproportionate ratio. Overall, our results exemplifies that the absolute Ne/N ratio should be used very carefully for conservation purposes, and particularly so in species with long generation time.
Evolution of genetic diversity through time
Both statistical methods utilized for inferring demographic history failed to detect the occurrence of a population bottleneck. Again, this may be explained by the sampling of only adult individuals. Under the assumption that the population size was dramatically reduced in the past 30 years, as surveys since the species’ recognition in 1942 have indicated, it could be too recent an event to detect the genetic consequences (Comité de rétablissement du Chevalier Cuivré 2004). Also, the heterozygosity excess test is sensitive to the severity of the population size reduction, and should detect only historical population bottlenecks that are severe (resulting census size < 40; Luikart & Cornuet 1998). Those methods have limitations, including the facts that mutation models are by no means perfect and the mutation rate estimate is ball-park at best. Yet, considering the facts that over 20 loci and a sample size of 165 were available for these analyses, and that using the SMM is statistically conservative according to other studies (Cornuet & Luikart 1996; Luikart & Cornuet 1998; Spencer et al. 2000), our main conclusions regarding the absence of a severe bottleneck in the copper redhorse appear justified.
Implications for conservation
The results of the simulations suggested that for a conservation plan for the copper redhorse to be successful from a genetic standpoint, one would need to maintain the effective population size to at least its current level. Thus, to achieve retention of 90% of the current diversity over the next 100 years, a commonly suggested conservation goal (Frankham et al. 2002), simulations predict a minimum effective population size of ∼400 is necessary. Assuming the average Ne/N ratio of 0.11 frequently observed in animals (Frankham 1995), this means that the recovery plan for copper redhorse should seek to maintain a census size of approximately 4000 adult individuals in order to maintain such a level of diversity in the long term. Given the current census size of the single known population approximated to 500 individuals only, the continuing decline of the copper redhorse, and the lack of recruitment, the continuation of a supportive breeding programme thus appears crucial in order to ensure the future of the population's genetic health, and should also include future monitoring of the genetic trends and consequences of conservation efforts (Bernatchez 2004; DeSalle & Amato 2004). In this context, the most positive asset of the copper redhorse's life cycle is its long generation time (and associated long lifespan and late maturation) which allows maintaining a high level of genetic diversity for a longer time period, and therefore allows more time to undertake proper conservation actions compared to species characterized by short generation time and lifespan.
Acknowledgements
This research was supported by the World Wildlife Fund Canada, the Ministère des Ressources Naturelles et de la Faune du Québec, the Fondation de la faune du Québec, and by the Canadian Research Chair in Conservation Genetics of Aquatic Resources. We are particularly grateful to Gérard Boucher, Alain Branchaud, Yves Chagnon, Jean Leclerc, Nathalie Vachon, Serge Pépin and Yves Gauthier for providing biological tissues, to Lucie Papillon and Robert Saint-Laurent for technical assistance in the laboratory, and to Anne-Marie Gale and Sean Rogers for helpful comments. We would also like to thank Jeff Haddrath and Michael Hansen for their precious help with DNA amplification of scales and bones, and particularly Robin S. Waples for his wise insights regarding the complexities of effective population size estimation and interpretation for long-lived species with overlapping generations.
References
This study represents part of C. Lippé's master research in the Bernatchez laboratory, a project that takes place into an important conservation plan aiming to preserve the endangered copper redhorse endemic to Québec, Canada. The global context of this study includes many other actions undertaken by the management committee, such as a supportive breeding programme, the construction of a migratory pass, and the creation of a sanctuary. All three authors share interests in conservation genetics.




