Phylogeography of postglacial range expansion in Nigronia serricornis Say (Megaloptera: Corydalidae)
Abstract
We have examined the effects of post-Wisconsinan glacial range expansion on the phylogeography of the saw-combed fishfly, Nigronia serricornis Say (Megaloptera: Corydalidae) because aquatic insects are under-represented in postglacial studies (and in phylogeography in general), and because the effects of ecological degradation on the population genetics of environmental indicator species like N. serricornis cannot be measured unless the underlying phylogeography is understood. Sequence data from a 630-base fragment of the mitochondrial cytochrome oxidase I (COI) gene were subjected to amova and nested clade analysis for 30 populations (n = 344) of N. serricornis. Both the amova and nested clade analysis revealed substantial population structure; 44.4% of the variance occured among populations. Three northward migrations are apparent: one from Tennessee into Illinois, Indiana, Wisconsin, Michigan and Ontario, a second that radiated eastward from Pennsylvania, and a third that moved along the coast from North Carolina into Connecticut, Massachusetts, Maine and then into New York. The latter two of these migrations were the result of contiguous range expansions, while the former expansion, out of Tennessee, appears to have been rapid with little gene flow from the source population. Additional clades included a group of haplotypes in central Kentucky that appear to have expanded along preglacial drainages, and clades in North Carolina and Georgia that have remained centrally located. Haplotype diversity decreased from south to north, a pattern that has been widely reported for animal and plant populations that expanded with the retreat of the Wisconsinan glaciation.
Introduction
Phylogeography, the combined analysis of gene genealogical data and geographic distribution, has become a powerful tool for inferring historical biogeographic events. Patterns of variation in mitochondrial and nuclear markers have allowed inference of past biogeographic events on every geographic scale from continental to local (Avise 2004). Despite recent progress in phylogeographic approaches, however, our understanding of historical biogeographic events remains incomplete due to large gaps in taxonomic and habitat sampling. Moreover, the use of phylogeographic insights to explain contemporary events such as changes in population structure due to environmental damage has just begun (Theodorakis 2003).
Aquatic insects, a group taxonomically under-represented in phylogeographic studies, are ideally suited for this type of research. Dispersal abilities vary enormously, from weakly dispersing stoneflies (e.g. Macneale et al. 2004), to wide-ranging dragonflies. Examples of such diverse abilities can even be seen within a single family, such as the water striders (Gerridae: Brinkhurst 1959; Zera 1981). Many aquatic insects share similar geographic distributions, allowing confirmation of inferred vicariance events using multiple species. The search for congruent patterns is also facilitated by the sheer number of aquatic insect species, conservatively estimated to be an order of magnitude greater than aquatic vertebrates (e.g. over 1500 aquatic insect species to approximately 180 species of native fishes in Wisconsin; Hilsenhoff 1981; Wisconsin Department of Natural Resources 1999). Additionally, many aquatic insects are key bio-indicator species because of their short generation times and environmental sensitivity (e.g. Hilsenhoff 1987), and contemporary changes in their genetic population structure may forecast problems for other aquatic denizens. Research on the population structure of some aquatic insects has been conducted (e.g. Zera 1981; Bunn & Hughes 1997; Hogg et al. 2002; Hughes et al. 2002; Schultheis et al. 2002; Wishart & Hughes 2003); however, few have examined patterns across a wide geographic range (but see Kauwe et al. 2004 and Ross & Ricker 1971).
Here we report a phylogeographic analysis of Nigronia serricornis Say (Megaloptera: Corydalidae), the saw-combed fishfly, an aquatic insect whose predatory larvae are restricted to highly oxygenated streams and are intolerant of organic enrichment (Hilsenhoff 1987). This species, like most megalopterans, is known to be a poor flier (Petersen 1974; Daly et al. 1998). Heilveil (2004), using flight mill and free-flight measurements, confirmed a low propensity for long-distance flight in the species, suggesting that dispersal may be severely limited. The range of N. serricornis extends from Florida to Ontario, east of the US Rocky Mountains, encompassing areas that were, during the last Pleistocene glacial event, both glaciated and unglaciated. Because the species is sensitive to organic enrichment and has limited dispersal capabilities, it is potentially a good model for future work examining anthropogenic impacts on underlying natural population genetic structure. To determine the basic population structure of N. serricornis and lay the groundwork for future work on anthropogenic effects, conventional population genetics analyses and nested clade analysis (NCA; Templeton 1987; 1998, 2001, 2004) were applied to cytochrome oxidase I (COI) sequences. NCA was appropriate for our data because it facilitates genetic analysis of individual clades and geographic regions that may have had very different and complex histories, as has been observed for other eastern North American organisms (e.g. Zamudio & Savage 2003).
The effects of historical events are important in eastern North America, where a series of Pleistocene glacial events transformed the landscape and caused mass migration and subdivision of species (e.g. Holman & Grady 1987; Leonard et al. 2000), leaving lasting effects on the genetic structure of both plants (e.g. Palme et al. 2003) and animals (e.g. Phillips et al. 2000). At its maximum (Fig. 1), the most recent (Wisconsinan) glaciation covered much of eastern North America from Illinois to New Jersey and north into Canada. Several biogeographic glacial effects have been extensively corroborated (Avise 2004). Many species show less variation in northern, formerly glaciated, areas than in unglaciated areas (e.g. Highton & Webster 1976; Sage & Wolff 1986; Armbruster et al. 1998), attributed to rapid postglacial population expansion (Hewitt 2001). Yet another effect of glaciation is on coastal migration corridors, which can change due to fluctuating sea levels as glaciers take up and release water; such changes can shape population structure by connecting previously isolated regions, e.g. use of the Gulf Coast dispersal corridor by terrestrial vertebrates (O’Brien et al. 1990; Ellsworth et al. 1994), and/or by isolating regions (e.g. Avise 1992).
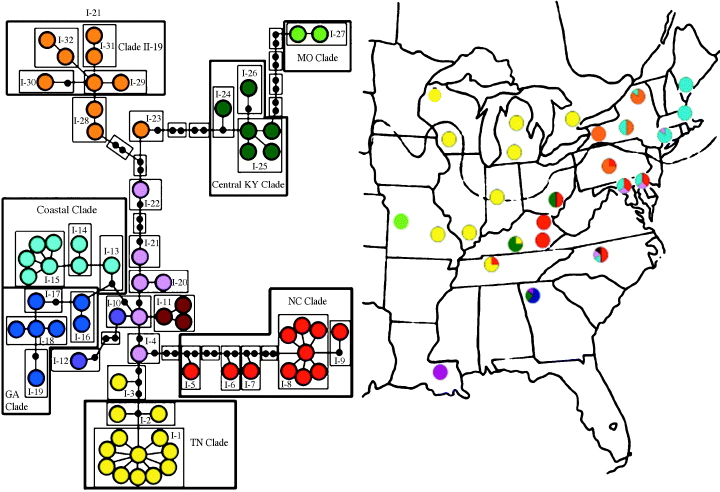
Sampling scheme and overlay of maximum-parsimony haplotype network on sampling geography. The populations on the map are colour-coded corresponding to the haplotype network to the left. The grey line indicates the approximate maximum advance of the Wisconsinan glaciation (c. 12 000 years ago). Modified from Fulton (1989). Note the two points of secondary contact in New York (the central and northeastern populations).
Some proposed glacial biogeographic effects have received less support from phylogeographic studies. The major indentation in the Wisconsinan glacial margin in southwestern Wisconsin and northwestern Illinois referred to as the ‘Driftless Area’ (Hartley 1966; Jacobs et al. 1997; but see Siegel & Mandle 1983) may possibly have served as a northern refugium for some populations (e.g. Chapco & Litzenberger 2002; Rowe et al. 2004), but the number of studies supporting this refugium is small.
Theodorakis (2003) laid out a set of criteria required to show causality between environmental stressors and subsequent changes to population genetic structure. A model organism for testing these criteria needs to be sensitive to anthropogenic change and have populations near developing areas with large enough genetic diversity that changes will be readily apparent. The organism should also be a poor disperser with low to moderate population densities, as it will be more susceptible to isolation and fragmentation than strong dispersers or organisms with large populations. Most relevant to this study, a model organism must be one in which the ‘temporality of effects’ (sensu Theodorakis 2003) can be determined. It must be possible to distinguish patterns due to older biogeographic events from those due to recent anthropogenic changes; in other words, a phylogeographic study of undisturbed populations is an absolute prerequisite for studying the population structure of disturbed populations. N. serricornis satisfies most of the above qualifications, with only the last remaining untested.
The general goals of this study are to better evaluate what aquatic insects can reveal about Pleistocene events, and to obtain a better understanding of the role of phylogeography in the population genetics of an environmental indicator species. We test the specific predictions that the relatively limited dispersal ability of N. serricornis will lead to a general pattern of significant genetic divergence among populations, and that in agreement with previous studies of a variety of species, haplotype diversity will decrease from south to north.
Materials and methods
Geography and collection of sample populations
To obtain a comprehensive understanding of the underlying historical population structure of Nigronia serricornis, 344 individual larvae representing 30 populations were sampled from across the range of the species (Fig. 1). The larvae of N. serricornis exhibit unambiguous morphological differences from the only other species in the genus, Nigronia fasciatus, allowing specific identification in the field. At each site, up to 18 N. serricornis larvae were collected by kick-netting and immediately preserved either on dry ice or in liquid nitrogen. Larvae were stored at −80 °C until DNA could be extracted. Whenever possible, sampling sites were selected where N. serricornis had recently (= 5 years) been collected by academic researchers or governmental bio-assessment units. One Corydalus cornutus (Vermillion County, Illinois) and four N. fasciatus larvae (representing 2 sites in West Virginia and 1 in Indiana) were collected and used as outgroup taxa (geographic sampling information available from the authors upon request). Voucher specimens for all Nigronia populations analysed were deposited in the insect collection of the Illinois Natural History Survey (INHS catalogue nos 39226 –39569 and 39570–39573, for N. serricornis and N. fasciatus, respectively).
MtDNA data collection
Initially, DNA was extracted from head, thoracic, and abdominal preparations. Extractions from the larval head capsule were used for later amplification, because they resulted in the cleanest extractions. All DNA extractions were performed using QIAGEN DNeasy™ kits, following the manufacturer's protocol for animal tissue samples. To offset the effects of an unknown polymerase chain reaction (PCR) inhibitor that co-eluted with the DNA, DNA elutions were diluted 1:150 with ultrapure water to ensure reliable amplification.
For all specimens, a 710-base fragment in a coding region of cytochrome oxidase subunit I (COI), previously shown to be useful in answering population-level questions (Wishart & Hughes 2003), was amplified using the primers LCO1490 (5′-GGTCAACAAATCATAAAGATATTGG-3′) and HCO2198 (5′-TAAACTTCAGGGTGACCAAAAAATCA-3′) of Folmer et al. (1994). Amplification reactions were prepared using the following protocol: 10.3 µL double processed tissue culture water (Sigma), 2.5 µL PCR Gold buffer (ABI), 1.0 µL 25 mm MgCl2, 1.0 µL 5 mm dNTPs, 1.0 µL each 20 mm primer, 0.2 µL AmpliTaq® Gold enzyme, and 8 µL diluted template. Amplification reactions were performed on an MJR PTC100, following the thermocycler protocol of Wishart & Hughes (2003). Amplification reactions were electrophoresed on 1.5% agarose gels for 45 min at 120 V, stained with ethidium bromide, and gels were visualized using UV light. PCR products were purified using QIAGEN QuickSpin™ kits as per the manufacturer's specifications.
The following sequencing reaction was prepared using the purified DNA: 5.2 µL 12.5% glycerin, 2 µL 5X sequencing buffer (ABI), 2 µL 20 mm PCR primer, 1.0 µL BigDye terminator (version 3.0, Applied Biosystems), and 6–8 µL purified DNA and subjected to 35 cycles of PCR using the conditions of Collins (2003). DNA fragments were sequenced in both directions. DNA sequencing was performed on an ABI 3730X1 by the University of Illinois Biotechnology Center.
Sequence verification and Bayesian phylogeny
Sequence chromatograms were verified and sequences were manually aligned, as no gaps were present in the sequences. All DNA sequences were deposited in GenBank (accession nos AY750172–AY750519). A 630-base fragment of COI was used in subsequent analyses. To ensure that PCR products were from true mitochondrial genes, a nucleotide-nucleotide blast search was performed on the most common haplotype. The corrected pairwise COI sequence divergence was calculated in paup* (version 4.0b10, Swofford 2003) using the same model as the Bayesian analysis (see below). A Bayesian phylogeny was constructed for unique COI haplotypes to identify ancestral haplotypes with the program mrbayes (version 3.0, Ronquist & Huelsenbeck 2003) using 2 000 000 trees, sampling 1/100 trees, with a burn-in value of 837, corresponding to generation 83 600. C. cornutus served as the outgroup taxon. The program modeltest (version 3.5, Posada & Crandall 1998) was used to determine the appropriate substitution model for COI (model = TVM + I + G). The burn-in value was selected by plotting the likelihood values a posteriori and selecting a generation beyond the point of stationarity (Ronquist et al. 2005).
Population genetic analyses
Standard population genetic and statistical analyses were employed to describe the main features of the data. In some of the population analyses, only samples with n = 8 (henceforth the ‘large’ samples) were included; a compromise between the desire to maximize samples, while including as much of the geographic range as possible (mean n of large populations = 16). As a measure of whether the data were consistent with selective neutrality, Tajima's D was calculated using arlequin 3.0 (Excoffier et al. 2005) for both the entire set of sequences considered together and for the individual large populations. The overall magnitude of population differentiation was measured by calculating FST across ungrouped samples with haplotype frequency data using arlequin (Excoffier et al. 2005). As an overall assessment of whether the population differentiation showed geographic structure, a comparison of the two fixation indices, GST (Nei 1982) and NST (Lynch & Crease 1990) was carried out using dnasp (version 4.0, Rozas & Rozas 1995). If the value of NST (analogous to GST, except that sequence differences as well as frequency differences between alleles are incorporated in the statistic) is larger than the value of GST for a given set of data, the presence of phylogeographic structure can be assumed (Pons & Petit 1996). Finally, the relationship between latitude and both the proportion of haplotypes at a sampling site (# haplotypes/n) and haplotype diversity were examined by linear regression, after dnasp was used to determine the haplotype diversity.
Phylogeographic analysis
Two methods, NCA and analysis of molecular variation (amova; Excoffier et al. 1992), were employed to examine the relationship between intraspecific phylogeny and geography. To implement the NCA, an explicit parsimony (Templeton et al. 1992) haplotype network was first constructed for the COI sequences using the program tcs (version 1.18, Clement et al. 2000), with connections made at the 95% parsimony limit (10 steps). The few ambiguous connections in the resulting haplotype network were resolved as per Crandall & Templeton (1993). The haplotype network was then overlain upon geographic space to look for gross patterns in the data, prior to being partitioned into nesting clades as per Templeton (1998). Information regarding clades that contained appropriate amounts of genetic and geographic variation was input into geodis (version 2.0, Posada et al. 2000) together with geographic sampling information, using the latitude/longitude method, to perform permutation tests for association between phylogeny and geographic distribution. Templeton's revised (2004) inference key was then applied to the results from geodis to determine the outcome of the NCA.
An amova was performed on populations grouped phylogenetically (groupings available upon request of the authors) using arlequin 3.0 (Excoffier et al. 2005). A global amova was carried out on the large population samples, using the following groupings of populations suggested by the NCA: clade II-19, Tennessee clade, Coastal clade, North Carolina clade, Georgia clade, and central Kentucky clade (see Fig. 1).
Results
Sequence variation and Bayesian phylogeny
The 630-base fragment of COI (containing 58 polymorphic and 38 parsimony-informative sites) was amplified and sequenced for 344 Nigronia serricornis larvae. Of these sequences, 68 unique haplotypes were recovered (see Table 1 for geographic occurrences). Approximately 94% of the polymorphic sites represented changes at third codon positions, and all substitutions were synonymous.
| Haplotype | GenBank Accession no. | n | Geographic occurrence |
|---|---|---|---|
| 1 | AY750172–AY750261 | 90 | Robertson County, TN (2); Jackson County (16), Washburn County (18) WI; Crawford County (15), Roscommon County (10), Calhoun County (17), Kalamazoo County (11), MI; Adair County KY (1) |
| 2 | AY750262 | 1 | Robertson County, TN (1) |
| 3 | AY750263 | 1 | Robertson County, TN (1) |
| 4 | AY750264–AY750265 | 2 | Robertson County, TN (2) |
| 5 | AY750266–AY750274 | 9 | Montgomery County, IN (9) |
| 6 | AY750275–AY750278 | 4 | Kalamazoo County, MI (4) |
| 7 | AY750279–AY750283 | 5 | Roscommon County, MI (5) |
| 8 | AY750284 | 1 | Crawford County, MI (1) |
| 9 | AY750285 | 1 | Crawford County, MI (1) |
| 10 | AY750286–AY750292 | 7 | Victoria County, Ontario (7) |
| 11 | AY750293–AY750299 | 7 | Robertson County, TN (7) |
| 12 | AY750300–AY750302 | 3 | Robertson County, TN (1) Pope County, IL (2) |
| 13 | AY750303–AY750312 | 10 | Ste. Genevieve County, MO (8), Union County, IL (2) |
| 14 | AY750313 | 1 | Montgomery County, NC (1) |
| 15 | AY750314 | 1 | Macon County, NC (1) |
| 16 | AY750315–AY750316 | 2 | Montgomery County, NC (2) |
| 17 | AY750317–AY750319 | 3 | Montgomery County, NC (3) |
| 18 | AY750320–AY75033, AY750354 | 19 | Montgomery County, NC (2); Robertson County, TN (1); Greenup County (6), Letcher County (2), KY; Cecil County (1), Montgomery County (7), MD |
| 19 | AY705338–AY750342 | 5 | Huntingdon County, PA (1); Cecil County (2), Montgomery County (2), MD |
| 20 | AY750343 | 1 | Hocking County, OH (1) |
| 21 | AY750344–AY750353 | 10 | Greenup County, KY (10) |
| 22 | AY750355 | 1 | Letcher County, KY (1) |
| 23 | AY750356 | 1 | Letcher County, KY (1) |
| 24 | AY750357 | 1 | Letcher County, KY (1) |
| 25 | AY750358 | 1 | Montgomery County, NC (1) |
| 26 | AY750359–AY750362 | 4 | Montgomery County, NC (3); Chattooga County, GA (1) |
| 27 | AY750363–AY750364 | 2 | Montgomery County, NC (2) |
| 28 | AY750365 | 1 | Montgomery County, NC (1) |
| 29 | AY750366 | 1 | Montgomery County, NC (1) |
| 30 | AY750367 | 1 | Chattooga County, GA (1) |
| 31 | AY750368 | 1 | Washington County, LA (1) |
| 32 | AY750369 | 1 | Fairfield County, CT (1) |
| 33 | AY750370–AY750386 | 17 | Cumberland County, ME (3), Tomkins County (2), Warren County (1), NY; Montgomery County (5), Cecil County (1), MD; Fairfield County, CT (5) |
| 34 | AY750387 | 1 | Montgomery County, NC (1) |
| 35 | AY750388–AY750409 | 22 | Cumberland County, ME (4); Fairfield County, CT (3); Norfolk County, MA (15) |
| 36 | AY750410–AY750418 | 9 | Cumberland County, ME (5); Fairfield County, CT (4) |
| 37 | AY750419–AY750421 | 3 | Cumberland County, ME (3) |
| 38 | AY750422–AY750423 | 2 | Norfolk County, MA (2) |
| 39 | AY750424 | 1 | Fairfield County, CT (1) |
| 40 | AY750425 | 1 | Chattooga County, GA (1) |
| 41 | AY750426–AY750427 | 2 | Chattooga County, GA (2) |
| 42 | AY750428 | 1 | Chattooga County, GA (1) |
| 43 | AY750429–AY750433 | 5 | Chattooga County, GA (5) |
| 44 | AY750434 | 1 | Chattooga County, GA (1) |
| 45 | AY750435 | 1 | Chattooga County, GA (1) |
| 46 | AY750436 | 1 | Chattooga County, GA (1) |
| 47 | AY750437–AY750440 | 4 | Montgomery County (3) Cecil County (1), MD |
| 48 | AY750441 | 1 | Cecil County, MD (1) |
| 49 | AY750442–AY750445 | 4 | Fairfield County, CT (3); Cecil County MD (1) |
| 50 | AY750446 | 1 | Warren County, NY (1) |
| 51 | AY750447 | 1 | Warren County, NY (1) |
| 52 | AY750448–AY750450 | 3 | Tomkins County (1), Warren County (1), NY; Huntingdon County, PA (1) |
| 53 | AY750451–AY750455 | 5 | Warren County (2), Erie County (1) NY; Huntingdon County, PA (2) |
| 54 | AY750456 | 1 | Erie County, NY (1) |
| 55 | AY750457–AY750458 | 2 | Erie County, NY (2) |
| 56 | AY750459–AY750478 | 20 | Huntingdon County, PA (11) Erie County (4), Warren County (5), NY |
| 57 | AY750479 | 1 | Warren County, NY (1) |
| 58 | AY750480 | 1 | Erie County, NY (1) |
| 59 | AY750481–AY750486 | 6 | Huntingdon County, PA (1); Erie County (4), Tomkins County (1), NY |
| 60 | AY750487 | 1 | Warren County, NY (1) |
| 61 | AY750488–AY750489 | 2 | Chattooga County, GA (1); Adair County, KY (1) |
| 62 | AY750490–AY750499 | 10 | Adair County, KY (8); Hocking County, OH (2) |
| 63 | AY750500–AY750501 | 2 | Adair County, KY (2) |
| 64 | AY750502–AY750503 | 2 | Adair County, KY (2) |
| 65 | AY750504–AY750507 | 4 | Adair County, KY (3); Warren County, NY (1) |
| 66 | AY750508 | 1 | Adair County, KY (1) |
| 67 | AY750509–AY750514 | 6 | Benton County, MO (6) |
| 68 | AY750515 | 1 | Benton County, MO (1) |
No stop codons were present in the mtDNA sequences, indicating that a true mitochondrial gene was amplified, as opposed to a nuclear pseudogene. The nucleotide-nucleotide blast search recovered a number of significant matches for COI sequences from other insects with over 80% sequence identity. Nigronia serricornis was different from Nigronia fasciatus by only a single amino acid, and the N. fasciatus sequences formed a monophyletic sister group to N. serricornis. Sequence divergence within N. serricornis ranged between 0 and 0.044 substitutions/site, with a mean divergence of 0.020 substitutions/site. The level of divergence between N. serricornis and N. fasciatus ranged between 0.195 and 0.245 substitutions/site, with a mean sequence divergence of 0.219 substitutions/site.
The Bayesian tree confirmed the monophyly of N. serricornis with respect to N. fasciatus, and posterior probabilities were greater than 90% for a majority of uncollapsed nodes (Fig. 2). The phylogeny rooted with Corydalus cornutus showed a group of haplotypes concentrated in Pennsylvania and New York (haplotypes 53–59; Fig. 2) to be ancestral for N. serricornis, with an overall ln(likelihood) of 2602.41.
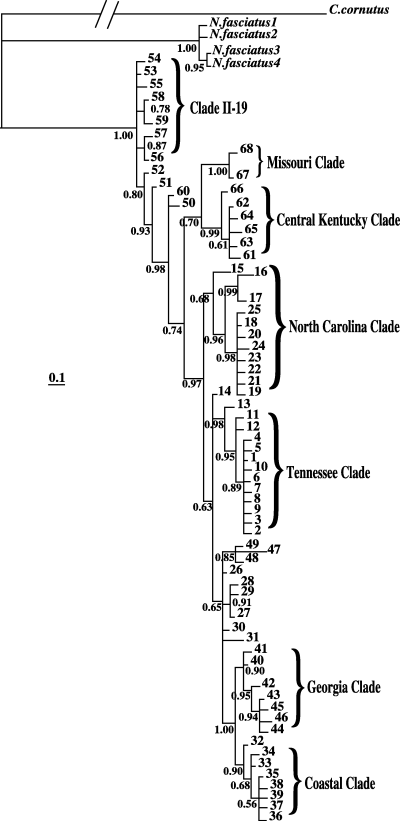
Bayesian phylogram based on COI sequences for Nigronia serricornis, Nigronia fasciatus, and Corydalus cornutus with C. cornutus as the outgroup taxon. The phylogenetic distance between the C. cornutus and the other species has been truncated. Posterior probabilities are given at each node. Numbers refer to the specific COI haplotype and are consistent with both the haplotype network (Fig. 5) and Table 1. For reference, the names of corresponding NCA clades are given, although it should be noted that ‘clade II-19’ is not a clade in the strict sense.
Population genetic analyses
Nucleotide diversity over the entire data set was 0.015 ± 0.008. The value of Tajima's D using all 344 sequences was −0.17524, not significantly different from 0 (P = 0.51), suggesting that no major historical selective events had occurred. Similarly, none of the Ds for the individual large populations were significantly different from 0. The global FST for the large population samples was very large (0.444), with 44.41% of the variance occurring between populations and 55.59% within populations. A comparison of the fixation indices NST and GST revealed a strong relationship between phylogeny and geography, with NST being much larger than GST (0.743 and 0.427, respectively). The regressions of haplotype proportion (Fig. 3a) and haplotype diversity (Fig. 3b) with sampling latitude were both significant (R2 = 0.475, P = 0.002; R2 = 0.388, P = 0.006, respectively) giving additional support for the importance of phylogeographic structure in the species. When a single outlier was removed (corresponding to a northern site of secondary contact in New York, see Discussion), more of the variation could be accounted for by latitude alone (R2 = 0.752, P < 0.001; R2 = 0.483, P = 0.002; for haplotype proportion and haplotype diversity, respectively).
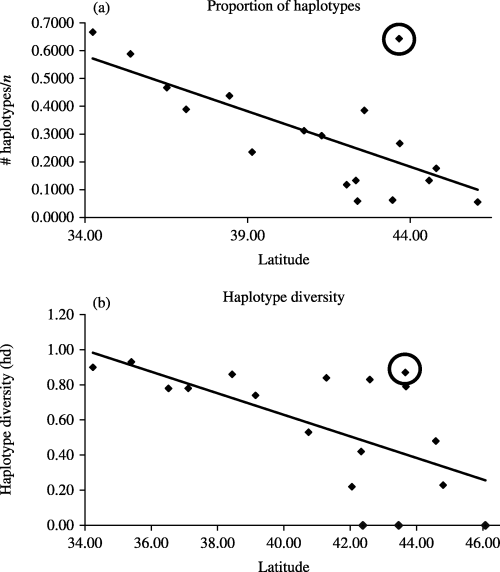
(a) Proportion of haplotypes vs. latitude for Nigronia serricornis. Data points represent all populations for which N = 15. The line of best fit is: y = −0.0399x+ 1.9382 and R2 = 0.475. The circled point represents a population in northeastern New York where secondary contact is occurring between two clades. (b) Haplotype diversity vs. latitude for N. serricornis. Data points represent all populations for which N = 15. The line of best fit is: y = −0.0613x+ 3.0798 and R2 = 0.3881. The circled point represents the same population described in (a).
Phylogeographic analyses
All of the haplotypes recovered from N. serricornis were successfully connected in a single network at the 95% confidence level by tcs. The tcs analysis, however, placed haplotype ‘1’ as the most ancestral, based on its frequency in the overall data set and its range of geographic distribution, but as described above, the Bayesian phylogenetic analysis placed the root with a group of haplotypes in Pennsylvania and New York. A Bayesian analysis rooted with haplotype ‘1’ had a ln(likelihood) of 2370.51, much lower than the 2602.41 for the Bayesian tree rooted with C. cornutus. The haplotype network was therefore polarized for the NCA using the results from the more likely Bayesian tree rather than the tca analysis. The haplotype network was partitioned into 32 one-step clades (Fig. 4), 19 two-step clades, 9 three-step clades, 4 four-step clades, and 2 five-step clades (Fig. 5). Of these 66 clades, 13 exhibited a significant correlation between geography and phylogenetic structure in the NCA. The specific outcomes from the NCA inference key can be seen in Table 2.
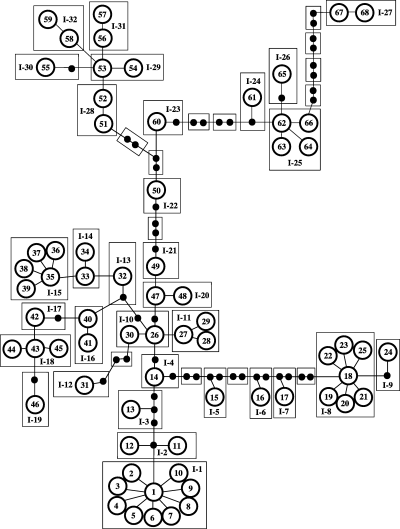
Nesting scheme for one-step clades. Circles represent unique COI haplotypes recovered from Nigronia serricornis, with arbitrary numbers that are consistent with Table 2. Boxes delineate individual one-step clades, with roman numerals representing the nesting size and arabic numerals representing the clade number.
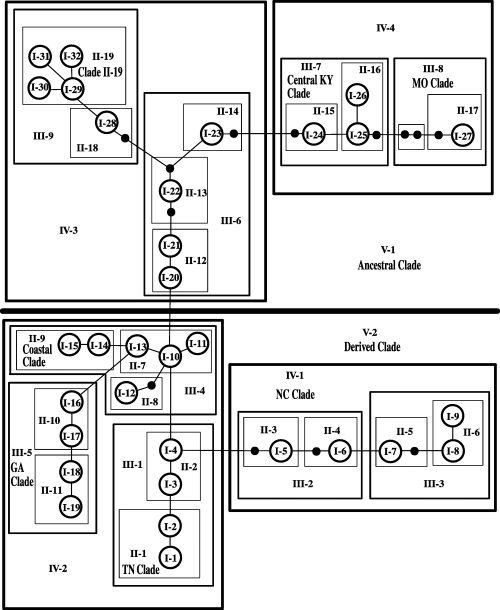
Nesting scheme for two-step, three-step, four-step, and five-step clades. Boxes delineate individual clades. Roman numerals represent the nesting size and arabic numerals represent the clade number.
| Clade | χ2 | Probability | Inference chain | Inferred event |
|---|---|---|---|---|
| I-1 | 396.9116 | 0.0000 | 1-2-3-5-6-13 — Yes | Past fragmentation followed by range expansion |
| I-15 | 24.4088 | 0.0000 | 1-2-11-12 — No | Contiguous range expansion |
| II-1 | 82.3738 | 0.0000 | 1-2-11-12-13 — Yes | Past fragmentation followed by range expansion |
| II-9 | 30.1236 | 0.0000 | 1-2-3-4 — No | Restricted gene flow with isolation by distance |
| III-1 | 142.0000 | 0.0000 | 1-2-11-12-13 — Yes | Past fragmentation followed by range expansion |
| III-4 | 117.9321 | 0.0000 | 1-2-3-5-6-7 — Yes | Restricted gene flow/dispersal |
| III-6 | 11.0000 | 0.0130 | 1-2-3-4-9 — No | Allopatric fragmentation |
| IV-1 | 20.8929 | 0.0330 | 1-2-11-17-4 — No | Restricted gene flow with isolation by distance |
| IV-2 | 398.5170 | 0.0000 | 1-2-3-4 — No | Restricted gene flow with isolation by distance |
| IV-3 | 41.1477 | 0.0000 | 1-2-11-12 — No | Contiguous range expansion |
| IV-4 | 28.0000 | 0.0000 | 1-19-20-2-11-12–13 — Yes | Past fragmentation followed by range expansion |
| V-1 | 200.2163 | 0.0000 | 1-2-3-4 — No | Restricted gene flow with isolation by distance |
| V-2 | 74.9657 | 0.0000 | 1-2-11-12 — No | Contiguous range expansion |
| Total | 276.6337 | 0.0000 | 1-2-11-12 — No | Contiguous range expansion |
Overlaying the un-nested haplotype network on the sampling geography yielded a close match between the two (Fig. 1). Six major clades were apparent: one covering Tennessee, Illinois, Indiana, Michigan, Wisconsin, and Ontario; a second centred in central Kentucky with haplotypes also occurring in southeastern Ohio and in Georgia; a third radiating out from North Carolina into Pennsylvania, southeastern Ohio, and eastern Kentucky; a fourth, only represented in Georgia, a fifth clade, containing the ancestral haplotypes, spanning across Pennsylvania and into New York; and the last clade occurring along the eastern seaboard, from North Carolina up to Connecticut and Massachusetts, and into Maine.
The results from the global amova are presented Table 3. All FST values were highly significant, with P values < 0.001 being returned by the arlequin permutation tests. Of most significance for historical interpretation, almost 20% of the variation occurs among clades.
| Source of variation | d.f. | Sum of squares | Variance components | Percentage of variation |
|---|---|---|---|---|
| Among groups | 5 | 32.960 | 0.09543 Va | 19.95 |
| Among populations /within groups | 14 | 30.559 | 0.12903 Vb | 26.97 |
| Within populations | 285 | 72.370 | 0.25393 Vc | 53.08 |
| Total | 304 | 135.889 | 0.47839 | |
| Fixation Indices | ||||
| F SC | 0.33693 | |||
| F ST | 0.46920 | |||
| F CT | 0.19948 | |||
Discussion
Populations of Nigronia serricornis displayed highly significant (FST = 0.444) large-scale biogeographic patterns that are consistent with many animal populations found in the eastern USA. As predicted by Hewitt (2000), there was decreased genetic diversity in glaciated areas as compared to populations from unglaciated areas. The N. serricornis results suggest that aquatic insects may have rich phylogeographic structures that can contribute to a better understanding of North American glacial biogeography. These results also indicate that sufficient genetic structure exists to allow older biogeographic events to be separated from more recent events. Therefore, the temporality of contemporary effects (sensu Theodorakis 2003) on N. serricornis population structure can be determined, making the species useful in future analyses of anthropogenic affects on aquatic indicator species. The phylogeographic patterns revealed are quite complex, as discussed below.
Intra- vs. interspecific divergence in Nigronia
The mean sequence divergence within N. serricornis (0.02 substitution/site) is an order of magnitude smaller than it is between N. serricornis and Nigronia fasciatus (0.219 substitution/site), the only other species in the genus Nigronia. The degree of sequence divergence within N. serricornis and between the two species of Nigronia is consistent with COI sequence divergences from other orders of insects. Hebert et al. (2003a) found that 196 of 200 lepidopteran species they examined exhibited interspecific COI sequence divergences in excess of 3%, and in Coleoptera, the most closely related order for which COI sequence divergence has been examined, 93.2% of the congeneric species pairs (n = 891 pairs) displayed interspecific divergences in excess of 4% (Hebert et al. 2003b). Therefore, it is reasonable to assume that N. serricornis exists as a single species across its range, despite its limited dispersal ability (Heilveil 2004). The Bayesian analysis also supported the monophyly of N. serricornis with respect to N. fasciatus and did not recover any significant subdivisions within the species.
Hewitt (2000) states that ‘Rapid colonization by this leading edge model in any part of the world should produce areas with reduced genomic variability.’ As Hewitt predicted, haplotype diversity was reduced in recently deglaciated areas; however, latitude only explained 39–48% of the variation in haplotype diversity in our study. Other factors such as secondary contact clearly play an important role in shaping patterns of haplotype diversity, as almost twice as much variation could be explained by latitude when a single point of secondary contact was removed from the correlation.
The major clades of N. serricornis in eastern North America
According to the Bayesian analyses rooted with Corydalus cornutus, clade II-19 (containing haplotypes 53–59; Fig. 5), represents the most ancestral haplotypes found in N. serricornis. The populations from which these haplotypes were recovered occur in central Pennsylvania and New York, the former being south of the Wisconsinan glacial maximum (Fulton 1989), and therefore may have served as a refugium. Typically, geographically widespread high-frequency haplotypes are assumed to be ancestral (Crandall & Templeton 1993; Castelloe & Templeton 1994), implying that haplotype ‘1’ in clade I-1 (Fig. 5) was the most ancestral: it occurs in 90 of the 344 animals sequenced and covers approximately 1100 km of the range of N. serricornis. An examination of the geographic distribution of clade I-1 (Fig. 1, in yellow), however, reveals that the clade predominantly occurs in areas that were uninhabitable during much of the Wisconsinan glaciation and therefore evolved more recently. The ancestral-like characteristics of clade I-1 will be discussed below.
The largest nesting level of the haplotype network consists of two main clades, V-1 (‘Ancestral’) and V-2 (‘Derived’) (Fig. 5). The populations represented in the Ancestral clade (the most basal haplotypes) stretch from Pennsylvania, New York and Maryland across to central Kentucky and into Missouri, representing the initial colonization of the eastern USA by N. serricornis. The Appalachian Mountain range, which neatly separates the Ancestral clade into clades IV-4 and IV-3, is likely responsible for the ‘restricted gene flow’ inferred by the NCA.
Within clade IV-4, a combination of populations from clade III-7 (‘central Kentucky’) and clade III-8 (‘Missouri’), there is evidence for past fragmentation followed by range expansion. According to Templeton (2004), the past fragmentation is further supported by the ‘larger-than-average’ number of mutational steps between the Missouri haplotypes (which are at the limit of the 95% CI for being connected to the rest of the network) and the central Kentucky clade. A similar pattern of geographically disjunct haplotypes was observed in Ambystoma maculatum Shaw (Phillips 1994).
Clade IV-3, the oldest of the four-step clades, experienced a contiguous range expansion out of Pennsylvania and into Connecticut and Maryland, with continued gene flow between the populations. More recently, however, allopatric fragmentation has occurred between these regions (NCA results for clade III-6, Table 3), potentially due to the poor dispersal ability of the species.
The Derived clade represents a second major wave of colonization by N. serricornis. Based on the polarization of the haplotype network, haplotypes in this clade appear to have spread out from North Carolina northwards along the coast, southwestward to Georgia, and westward to Tennessee and into the northern Midwest. The populations in this clade have undergone contiguous range expansion, and it is probable that the western migration out of North Carolina skirted the southern edge of the Appalachians. The Derived clade breaks down into clades IV-1 and IV-2, both of which experienced restricted gene flow and isolation by distance. The former of these, clade IV-1 (‘North Carolina’), radiates out from North Carolina into Tennessee, Kentucky, Ohio, Pennsylvania and Maryland. Part of the populations in this clade reside in eastern Kentucky, across the Cumberland Mountains and the intervening lowlands, a probable cause of the restricted gene flow inferred for this clade.
Clade IV-2 on the other hand, covers a vast amount of geographic space from the East Coast south to Georgia and north into Michigan and Wisconsin. This clade displays some of the most interesting patterns observed in this study. Clade IV-2 is comprised of clades III-5 (‘Georgia’), III-4, and III-1. The Georgia clade is restricted in distribution to Georgia and displays no phylogeographic structure. In contrast, clades III-4 and III-1 show two strikingly different patterns. Within clade III-4, is a large group of haplotypes that are spread out along the East Coast (clade II-9, ‘Coastal’). The Coastal clade as a whole has experienced restricted gene flow, possibly influenced by changes in coastal habitat induced by the Wisconsinan retreat. As the Wisconsinan retreated, water was released into the ocean, moving the shoreline inland (Fulton 1989) and altering the connectivity between coastal populations of animals. Within the Coastal clade, clade I-15 has undergone contiguous range expansion. Gene flow within this clade has allowed for haplotype transfer between populations in Maryland, Connecticut, Massachusetts, New York, and Maine. Similar movement of haplotypes from the southeastern USA northward along the coast has been observed in A. maculatum (Zamudio & Savage 2003).
The phylogeographic patterns of clade II-1 (‘the Tennessee clade’) are driven by clade I-1. This clade experienced a past fragmentation coupled with a range expansion that began in Tennessee and pushed north into Illinois, Missouri, Indiana, Wisconsin, Michigan and Ontario (Fig. 1, pictured in yellow). Similar to the findings of Seddon et al. (2001) and Alexandrino et al. (2002), the NCA made no inferences about the rapidity at which the range expansion took place. When the haplotype network is overlain on the sampling geography, it becomes clear that the expansion occurred over a short geological time period. The paucity of unique haplotypes recovered from the northern populations in this clade supports a rapid rate of range expansion with little gene flow occurring between the newly established populations and those found further south.
The recent, rapid, range expansion of the Tennessee clade also explains why clade I-1 displayed basal-like characteristics, as previously mentioned. During a rapid range expansion with little to no gene flow between populations, newly colonized populations have a limited number of haplotypes from which to be founded, allowing a single haplotype to spread over a large geographic area. If the range expansion occurred recently, novel haplotypes that have evolved will not have dispersed far, and the number of populations sampled from within the boundaries of the range expansion will bias the haplotype frequency towards the single ‘colonizing’ haplotype. Additionally, few mutations will have had time to accrue in any single lineage. All of the haplotypes occurring in clade I-1 for N. serricornis are single-step mutations that, based on their geographic distribution, have arisen since the Wisconsinan glacial retreat. We are currently working on obtaining sequences for other genes to examine some migration patterns at a finer scale.
Only a single haplotype was recovered from either population in Wisconsin and it is of Tennessee origin. The lack of genetic diversity in Wisconsin populations, in spite of reasonable sample sizes (n = 16 and 18), suggests that N. serricornis did not use the Driftless Area as a glacial refugium during the Wisconsinan glaciation unlike chipmunks (Rowe et al. 2004). Rather, N. serricornis followed the south-to-north migration pattern seen for most other organisms examined (e.g. Edmands 2001; Brant & Orti 2003; Zamudio & Savage 2003).
As predicted, the life history traits of N. serricornis have also shaped the population structure in the species. Out of the 13 clades for which an association could be found between phylogeny and geography, 10 clades experienced either population fragmentation or restricted gene flow. This preponderance of fragmentation suggests that individuals migrate between populations only in extremely rare instances, providing further support for limited dispersal in the species.
Phylogeographic congruence with other species in eastern North America
The only other study of postglacial range expansion by eastern North American aquatic insects was performed by Ross & Ricker (1971) on the plecopteran genus Allocapnia. Using distributional information together with morphological variation, Ross and Ricker stated that ‘only three species of Allocapnia (granulata, ninipara, and rickeri) appear to have moved northward through [northern Illinois and Indiana].’ Ross and Ricker felt that the slow, sandy streams present in Illinois and Indiana served as a barrier to migration for clean-water (i.e. dissolved-oxygen sensitive) species, such as Allocapnia illinoisensis. Ross and Ricker postulated that A. illinoisensis colonized Wisconsin and Minnesota via New York and Ontario, based on a ‘slight’ morphological variation found in A. illinoisensis from two Illinois streams and distributional information. Insects were obtained by solicitation, so it is unclear how intensely streams in this area were sampled and whether a priori assumptions about distribution influenced collecting effort. Interestingly, Ross and Ricker suggested that A. rickeri, another clean-water species, migrated through northern Illinois and Indiana, even though the species exhibited an almost identical distribution to A. illinoisensis (Ross & Ricker 1971). The molecular data from the pollution-intolerant N. serricornis suggest that dissolved oxygen (resulting from low flow and high sedimentation) levels in Illinois and Indiana streams were not barriers to migration for this species. Instead, the Tennessee clade migrated through the area en route to Michigan and Wisconsin. The inferred pattern of migration from Michigan into Ontario is identical to that observed for the ‘interior clade’ of the spotted salamander (Zamudio & Savage 2003), further supporting migration through Illinois and Indiana.
By overlaying the haplotype network on the sampling geography, points of secondary contact between two population expansions in central and northeastern New York (Fig. 1) become apparent. In these populations, haplotypes from both clade II-19 and the Coastal clade are present. The re-integration of haplotypes from these two clades artificially inflates both the proportion of haplotypes and the haplotype diversity above that expected for a similarly located population for either clade alone (Fig. 3).
Masta et al. (2003) point out that the phylogeographic patterns seen in aquatic animals may not reflect those of terrestrial animals. The major reason for this is that the distribution of aquatic organisms is often tied to hydrologic connectivity, as shown for golden crayfish (Fetzner & Crandall 2003). A good example of this relationship can be seen in the distribution of the central Kentucky clade (Fig. 1, pictured in dark green). Haplotypes from this clade occur in central Kentucky, Georgia and southeastern Ohio. The clade is noticeably absent from eastern Kentucky in populations that lie on a straight path between the central Kentucky and southeastern Ohio sites. The NCA did not find any significant patterns in the distribution of the central Kentucky clade. One explanation for this visual pattern is that subsequent to the initial colonization of southeastern Ohio, the central Kentucky clade became locally extinct in the eastern portion of Kentucky (similar to events described in Masta et al. 2003). Taking the aquatic nature of N. serricornis into account, however, a more plausible explanation becomes apparent: that the expansion of the central Kentucky clade followed river channels. A comparison of the distribution of this clade to a map of preglacial drainages composed by Mayden (1988) supports the probability of a drainage-mediated range expansion (Fig. 6). The population of N. serricornis sampled in central Kentucky is located in the Green River drainage basin, which, prior to the Wisconsinan glaciation, was closer by river distance to the population in southeastern Ohio than to either of the populations in eastern Kentucky. Drainage systems may guide population expansions in aquatic animals complicating the relationship between haplotype diversity and latitude. One solution to this complication is to perform the NCA using river distances, as opposed to latitudinal distance (e.g. Fetzner & Crandall 2003). Fetzner & Crandall (2003) suggest that river distance is the appropriate distance measure to use for organisms with an aquatic dispersal stage. Although the use of river distance would allow a better explanation of events in the central Kentucky clade, overall, terrestrial distance is more appropriate for use with N. serricornis, as dispersal occurs primarily during its terrestrial adult stage, and work on a confamilial showed extremely limited larval movement (Hayashi & Nakane 1989), further supporting the use of terrestrial distance over river distance.
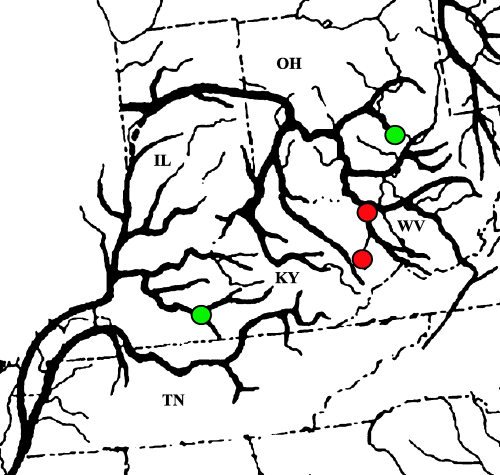
Overlay of the central Kentucky clade on preglacial drainages. The green circles represent populations where central Kentucky clade haplotypes occur, while the red circles represent populations where those haplotypes are absent. Note that even though these two red populations are closer to the central Kentucky population via linear distance, they are much further away than the Ohio population by river distance. Modified from Mayden et al. (1988).
Conclusions
Postglacial range expansions have clearly left their mark on contemporary populations of Nigronia serricornis. Ten of the 13 clades that were significantly correlated with geography exhibited either population fragmentation or restricted gene flow, supporting our prediction based on limited adult dispersal. Haplotype diversity is highest in the southern end of the range and is especially low in the northern Midwest as a result of a rapid range expansion, as seen for most organisms studied in this geographic region. Genetic diversity is slightly higher at specific latitudes along the East Coast where a contiguous range expansion allowed for the retention of genetic diversity, as compared to the northern Midwest. Below the extent of the Wisconsinan glacial maximum, diversity increases. There is no evidence for a northern glacial refugium in the Driftless Area of Wisconsin. Additionally, hot spots of genetic diversity can be found in New York and Ohio where secondary contact has been observed. Areas where two expanding clades have met should be good locations for subsequent studies of anthropogenic effects, not only because of the simple increase of alleles to use as genetic markers, but also because the mix of genotypes adapted to two different regions affords much opportunity for the action of selection.
Acknowledgements
The authors would like to gratefully thank Dr A. A. Valerio for his assistance with the Bayesian analyses performed in this study. Recently collected populations of Nigronia serricornis were identified by John Wuycheck (MI DEP), a number of US EPA bio-assessment units (New England, NY, PA, MD, WV, MO), Dr R. E. DeWalt (INHS), Dr K. Johnson (U Ohio), Dr S. Krauth (U Wisconsin), Dr D. Lenat (formerly of NC DEP), and Dr C. Pennuto (Buffalo State College). Additionally, Dr D. Lenat provided the samples from North Carolina and Dr C. Pennuto provided samples of the Ontario population. The following people generously donated their time while assisting in the field: R. Brinkman, T. Brown, A. Encalada, S. Fay, M. Goebel, Dr S. Hamilton, A. Heilveil, D. Heilveil, R. Houtman, E. Lacey, B. Newton, A. Peralta, R. Ruffing, Dr A. A. Valerio, and D. Wilkening. Valuable insights on molecular techniques and analysis were provided by Dr J. Anderson, L. Anderson, Dr S. Collins, L. Guest, Dr S. Lyons-Sobaski, H. Patch, Dr K. Ramsdell, K. Rowe, Dr A. A. Valerio, and K. Walden. Dr C. Phillips was helpful in interpreting the phylogeographic patterns. The authors would also like to thank J. Buhay, Dr S. Cameron, and three anonymous reviewers for comments on the manuscript. Funding for this research came from the North American Benthological Society Student Research Grant, the Harlie M and Frances M. Clark research support grant, the Illinois State Academy of Sciences Research Support Grant, and an On-Campus Dissertation Research Grant from the University of Illinois Graduate College.
References
J. S. Heilveil is a postdoctoral fellow at North Dakota State University. This study was part of his PhD research in Entomology at the University of Illinois at Urbana–Champaign. S. H. Berlocher is a professor in the Department of Entomology at the University of Illinois at Urbana–Champaign where he performs research on speciation, especially sympatric speciation.




