Lifespan extension by suppression of autophagy genes in Caenorhabditis elegans
Communicated by: Yoshimi Takai
Abstract
Lifespan is regulated by a complex combination of environmental and genetic factors. Autophagy, which is a bulk degradation system of macromolecules and organelles, has an important role in various biological events. In Caenorhabditis elegans, several autophagy genes have been shown to have a role in promoting longevity, but many other autophagy genes have not been examined for their role in the lifespan regulation. Here we have systematically examined the effect of RNAi suppression of 14 autophagy genes on lifespan. While maternal RNAi of autophagy genes in wild-type worms tended to reduce lifespan, maternal RNAi of each of seven autophagy genes in the insulin/IGF-1 receptor daf-2 mutants extended lifespan. Remarkably, RNAi of unc-51/atg-1, bec-1/atg-6 or atg-9, from young adult, i.e. after development, extended lifespan in both wild-type animals and daf-2 mutants, although RNAi of one or two genes shortened it. Moreover, our analysis suggests that the lifespan extension, which is induced by RNAi of unc-51, bec-1 or atg-9 after development, does not require the transcription factor daf-16, the NAD+-dependent protein deacetylase sir-2.1 or the genes related to mitochondrial functions. Collectively, our results suggest that autophagy may not always be beneficial to longevity, but may also function to restrict lifespan in C. elegans.
Introduction
Lifespan is regulated by both genetic and environmental factors (Kenyon 2005). It has been shown that the insulin/IGF-1 signaling pathway plays a pivotal role in lifespan regulation and that dietary restriction extends lifespan in many organisms. Moreover, many other factors have also been shown to regulate lifespan.
Autophagy is the major degradation pathway in eukaryotic cells, in which long-lived proteins and cytoplasmic organelles are sequestered within double membrane vesicles called autophagosome and delivered to lysosome, and then degraded and recycled (Yorimitsu & Klionsky 2005). Autophagy is evolutionarily conserved in eukaryotes from yeast to mammals and has important roles in various cellular functions.
In Caenorhabditis elegans, the insulin/IGF-1 receptor daf-2 mutants have been shown to have extended lifespan (Kenyon et al. 1993; Kimura et al. 1997). In the first report showing genetic evidence for the role of autophagy in longevity, maternal RNAi inactivation of an autophagy gene bec-1, the C. elegans ortholog of yeast ATG6 or human beclin 1, was shown to suppress the lifespan extension of daf-2 mutants and slightly shorten the lifespan of wild-type worms (Meléndez et al. 2003). Then, several reports appeared. In daf-2 mutants, RNAi of bec-1, atg-7, lgg-1, the C. elegans ortholog of yeast ATG8, atg-9 or lgg-3, the C. elegans ortholog of yeast ATG12, or mutations in bec-1 or atg-18 shortened lifespan (Hars et al. 2007; Hansen et al. 2008; Tóth et al. 2008). In several other long-lived mutants in C. elegans, for example, eat-2 mutants, which are a genetic model of dietary restriction, RNAi or mutations of several autophagy genes, such as bec-1, unc-51, the C. elegans ortholog of yeast ATG1, atg-7 or atg-18, shortened lifespan (Jia & Levine 2007; Hansen et al. 2008; Tavernarakis et al. 2008; Tóth et al. 2008). The lifespan-shortening effects of inactivation of autophagy genes on long-lived mutants are generally more significant than those on wild-type worms. While strong inactivation of some of autophagy genes might shorten lifespan by affecting development, it is generally believed that autophagy is beneficial to longevity in C. elegans. The beneficial effect of autophagy on longevity has also been shown in other organisms, although it should be noted that inhibition of autophagy genes does not always reduce normal lifespan in several organisms, including Drosophila (Cuervo 2008; Ren et al. 2009). It is also noted that in C. elegans, RNAi of bec-1 from young adult, i.e. after development, did not shorten lifespan in some of long-lived mutants, such as the mutants, in which translation is suppressed (Hansen et al. 2008).
As several autophagy genes should have functions outside of the autophagy pathway and also might play an important role in development, a comprehensive survey of all of autophagy genes in lifespan regulation should be required for assessing the role of autophagy in lifespan regulation. Then, we have systematically examined the effect of RNAi suppression of 14 autophagy genes in C. elegans on lifespan in this study. Our results show that although maternal RNAi of some of autophagy genes in wild-type worms reduced lifespan, in most other cases maternal RNAi or RNAi of autophagy genes after development did not reduce lifespan. Remarkably, RNAi of unc-51, bec-1 or atg-9 after development extended lifespan in wild-type animals and several other mutants. Thus, our results suggest that autophagy may not always be beneficial to longevity, but may also function to restrict lifespan in C. elegans.
Results
Searching for autophagy genes in C. elegans
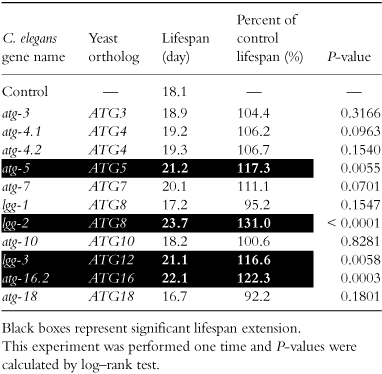
To systematically examine the effect of RNAi suppression of autophagy genes on lifespan in C. elegans, we searched the C. elegans genome for C. elegans autophagy genes. In WormBase, we identified 15 C. elegans orthologs of 12 yeast autophagy genes (Table 1); 10 C. elegans orthologs (unc-51 (yeast ATG1), atg-3 (yeast ATG3), atg-4.1 (yeast ATG4), atg-4.2 (yeast ATG4), bec-1 (yeast ATG6), atg-7 (yeast ATG7), lgg-1 (yeast ATG8), lgg-2 (yeast ATG8), atg-9 (yeast ATG9) and atg-18 (yeast ATG18)) were identified by using yeast autophagy genes, and 5 C. elegans orthologs (atg-5 (yeast ATG5), atg-10 (yeast ATG10), lgg-3 (yeast ATG12), atg-16.1 (yeast ATG16) and atg-16.2 (yeast ATG16)) were identified by using mammalian orthologs, which are listed in the reference (Klionsky et al. 2003). Two genes (atg-2/M03A8.2, the C. elegans ortholog of the human ortholog of yeast ATG2, and atg-13/D2007.5, the ortholog of human KIAA0652, which was identified as the ortholog of yeast ATG13 by PSI (Position-Specific Iterated)-BLAST search) were not listed in Table 1. Although C. elegans orthologs of ATG13, ATG14, ATG15, ATG16 and ATG17 were also reported to be identified recently (Khan et al. 2008), our search did not identify these genes as orthologs. Instead, as C. elegans orthologs of ATG16, our search using the mouse ortholog of yeast ATG16 identified different genes (atg-16.1/F02E8.5 and atg-16.2/K06A1.5). Thus, in this study, we performed RNAi suppression of 14 C. elegans autophagy genes in total, which are listed in Table 1 except for atg-16.1.
| Yeast ortholog | C. elegans sequence name | C. elegans gene name |
|---|---|---|
| ATG1 | Y60A3A.1 | unc-51 |
| ATG2 | — | — |
| ATG3 | Y55F3AM.4 | atg-3 |
| ATG4 | Y87G2A.3 | atg-4.1 |
| ATG4 | ZK792.8 | atg-4.2 |
| ATG5 | Y71G12B.12† | atg-5 |
| ATG6 | T19E7.3 | bec-1 |
| ATG7 | M7.5 | atg-7 |
| ATG8 | C32D5.9 | lgg-1 |
| ATG8 | ZK593.6 | lgg-2 |
| ATG9 | T22H9.2 | atg-9 |
| ATG10 | D2085.2‡ | atg-10 |
| ATG12 | B0336.8† | lgg-3 |
| ATG13 | — | — |
| ATG14 | — | — |
| ATG15 | — | — |
| ATG16 | F02E8.5‡ | atg-16.1 |
| ATG16 | K06A1.5‡ | atg-16.2 |
| ATG17 | — | — |
| ATG18 | F41E6.13 | atg-18 |
| ATG22 | — | — |
| ATG29 | — | — |
- † They were identified by the search using human orthologs of yeast autophagy genes.
- ‡ They were identified by the search using mouse orthologs of yeast autophagy genes.
Effect of RNAi of autophagy genes on lifespan
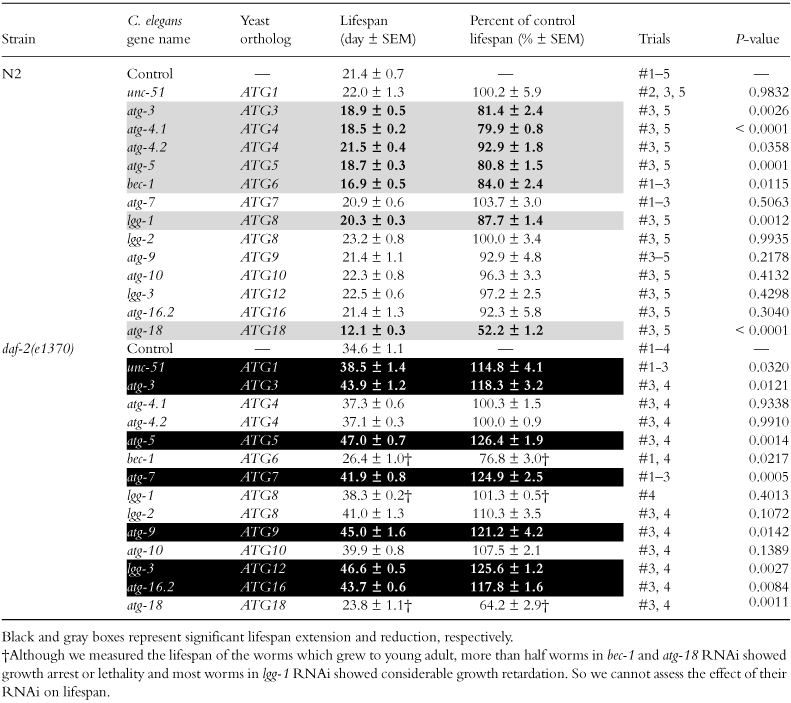
First, we performed maternal RNAi suppression of 14 autophagy genes in wild-type N2 worms. While maternal RNAi of 7 genes reduced lifespan in N2, maternal RNAi of the other seven genes did not affect lifespan significantly (Table 2, and Fig. 1a,b and c for unc-51, bec-1 and atg-9, respectively). Then, we performed maternal RNAi in a daf-2 mutant (daf-2(e1370)). Although maternal RNAi of two genes, bec-1 and atg-18, apparently reduced lifespan in the daf-2 mutant (Table 2), a large number of these RNAi-treated daf-2 worms showed developmental abnormalities (more than half worms in bec-1 and atg-18 RNAi), such as growth arrest and lethality. Also, most of lgg-1 RNAi-treated daf-2 worms showed considerable growth retardation, although they eventually grew to become young adults. Therefore, we cannot assess the effect of maternal RNAi suppression of these three genes on the lifespan of the daf-2 mutant. In the other 11 genes, maternal RNAi of them did not cause apparent developmental abnormalities. Surprisingly, maternal RNAi of seven genes out of the 11 genes extended lifespan by about 15%–25% in the daf-2 mutant (Table 2, and Fig. 1a and c for unc-51 and atg-9, respectively). Thus, our results show that while maternal RNAi of autophagy genes tends to reduce lifespan in wild-type worms, it does not reduce the lifespan of the daf-2 mutants but rather tends to extend their lifespan.
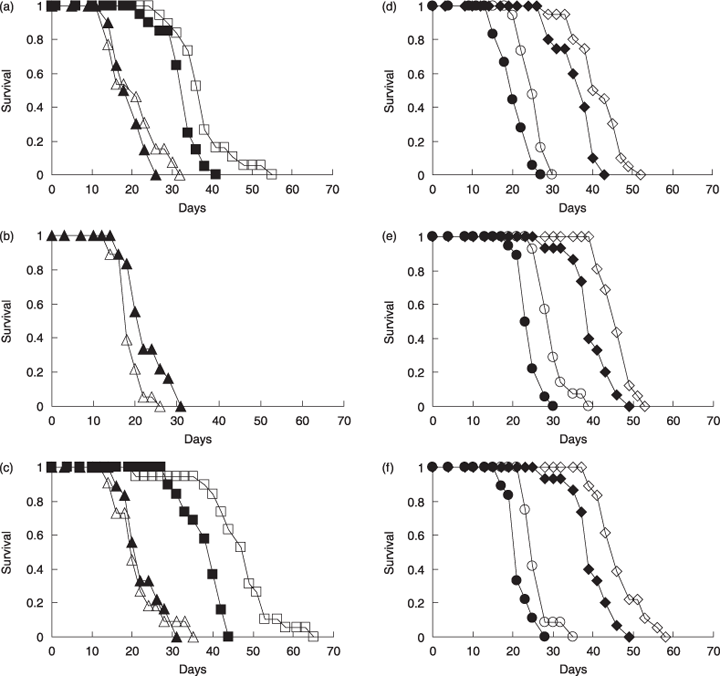
Lifespan in RNAi of unc-51/atg-1, bec-1/atg-6 and atg-9. Solid symbols indicate control and open symbols indicate RNAi of each gene. (a–c) Maternal RNAi of unc-51 (a), bec-1 (b) and atg-9 (c) in N2 (triangles) and in the daf-2(e1370) mutant (squares). The curves in RNAi of bec-1 in the daf-2 mutant were not shown because many worms in RNAi of bec-1 in the daf-2 mutant caused developmental abnormalities. (d–f) RNAi of unc-51 (d), bec-1 (e) and atg-9 (f) after development in N2 (circles) and in the daf-2(e1370) mutant (diamonds). Curves shown are representative of several measurements. Lifespan measurements were performed at 20 °C.
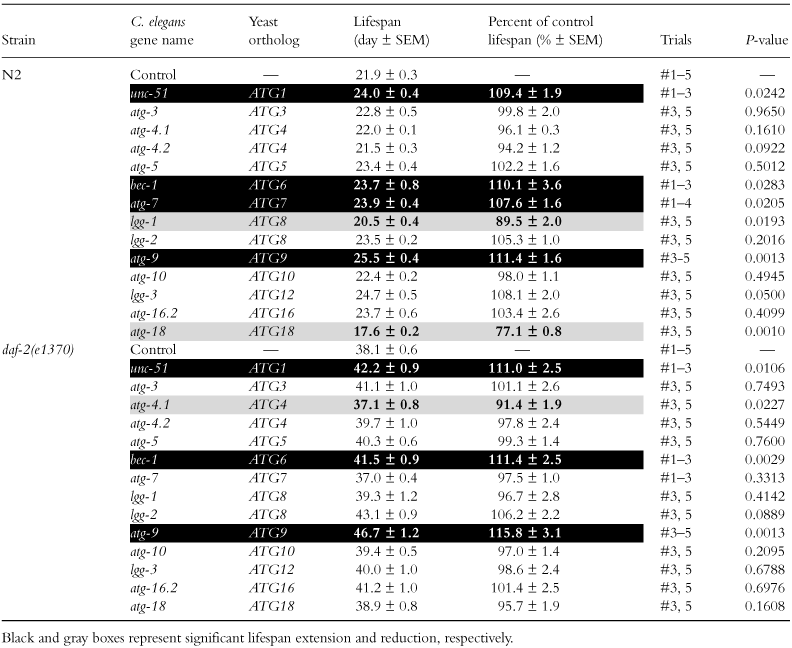
Next, to avoid developmental abnormalities, we performed RNAi suppression of 14 autophagy genes after development. Then, RNAi of four genes, unc-51, bec-1, atg-7 or atg-9, extended lifespan by about 10% in N2, and RNAi of three of these four genes except atg-7 also extended lifespan by about 10%–15% in the daf-2 mutant, although RNAi of two genes, lgg-1 or atg-18, in N2 and RNAi of one gene, atg-4.1, in the daf-2 mutant reduced lifespan (Table 3 and Fig. 1d,e and f for unc-51, bec-1 and atg-9, respectively). These results show that when autophagy genes are suppressed after development, suppression of autophagy genes seldom reduces lifespan, but often extends lifespan.
Suppression of autophagy genes can extend lifespan in daf-16 or sir-2.1 mutants
We then investigated whether the lifespan extension by suppression of autophagy genes is mediated through the known pathways that regulate lifespan in C. elegans. As RNAi of unc-51, bec-1 or atg-9 after development has been found to extend lifespan in both N2 and daf-2(e1370) worms, we performed RNAi of each of the three genes after development in several other mutants.
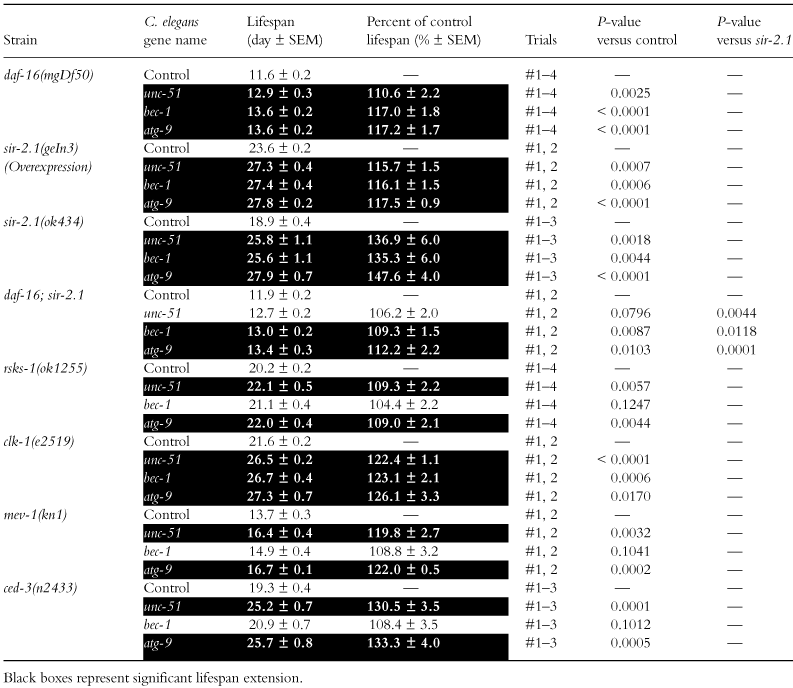
The lifespan extension in the reduction of DAF-2 function and the insulin/IGF-1 pathway requires the transcription factor DAF-16 (Lin et al. 1997; Ogg et al. 1997). So we examined whether suppression of autophagy genes extends the lifespan of a daf-16-deficient worm (daf-16(mgDf50)). Our lifespan measurements have shown that RNAi of the three autophagy genes, unc-51, bec-1 or atg-9, extended lifespan by about 10%–15% in the daf-16(mgDf50) mutant (Table 4 and Fig. 2a). Thus, it is likely that DAF-16 is dispensable for the lifespan extension by the autophagy gene suppression.
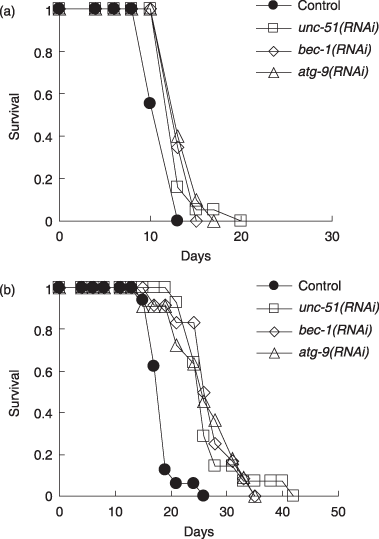
Lifespan in daf-16 and sir-2.1 mutants in RNAi after development. (a) The daf-16(mgDf50) mutant. (b) The sir-2.1(ok434) mutant. Solid circles indicate control, open squares indicate RNAi of unc-51, open diamonds indicate RNAi of bec-1, and open triangles indicate RNAi of atg-9. Curves shown are representative of several measurements. Lifespan measurements were performed at 20 °C.
SIR2, an NAD+-dependent protein deacetylase, is one of the major genes that regulate lifespan and is suggested to be involved in dietary restriction-induced longevity (Chen & Guarente 2007). Over-expression of sir-2.1, a C. elegans SIR2 gene, is known to extend lifespan, in a daf-16-dependent manner, and sir-2.1 deletion slightly reduces lifespan (Tissenbaum & Guarente 2001; Wang & Tissenbaum 2006). Our measurements show that RNAi of the three autophagy genes extends lifespan by about 15%–20% in a sir-2.1 over-expression mutant and by about 35%–50% in a sir-2.1-deficient mutant (sir-2.1(ok434)) (Table 4 and Fig. 2b). RNAi of four other autophagy genes was also shown to extend lifespan in the sir-2.1 mutant (Table 5). These results suggest that SIR-2.1 is not required for the autophagy gene suppression to extend lifespan.
As the extent of the lifespan extension by the autophagy gene suppression is larger in the sir-2.1 mutant, we performed RNAi suppression of autophagy genes in a sir-2.1 and daf-16 double mutant. The lifespan of the daf-16(mgDf50); sir-2.1(ok434) mutant was shorter than that of N2 or the sir-2.1 mutant, and roughly equal to that of the daf-16(mgDf50) mutant (Table 4). RNAi of bec-1 or atg-9 extended the lifespan of the daf-16(mgDf50); sir-2.1(ok434) mutant, but the extent of the lifespan extension was smaller than that in the sir-2.1 mutant (Table 4). This suggests that daf-16 play a role in the large lifespan extension by suppression of autophagy genes in the sir-2.1 mutant.
Suppression of autophagy genes can extend lifespan in other mutants that affect lifespan
Inactivation of TOR or inhibiting translation has been shown to extend lifespan in C. elegans (Vellai et al. 2003; Hansen et al. 2007; Pan et al. 2007; Syntichaki et al. 2007). TOR regulates protein synthesis and suppresses autophagy (Wullschleger et al. 2006). It has been reported that unc-51, bec-1 and atg-18 are required for the lifespan extension produced by inactivation of TOR, whereas bec-1 is not required for the lifespan extension produced by inhibiting translation (Pan et al. 2007; Hansen et al. 2008; Tóth et al. 2008). We asked whether suppression of autophagy genes extends the lifespan in the S6 kinase rsks-1 mutant, in which translation has been shown to be suppressed (Pan et al. 2007; Hansen et al. 2008). Our results show that RNAi of unc-51 or atg-9 extended the lifespan of the rsks-1(ok1255) mutant, although the extent was slightly less than in N2 (Table 4). Thus, RSKS-1 may be dispensable for the autophagy gene suppression-induced lifespan extension.
Many of genes that are involved in mitochondrial functions are known to influence lifespan (Feng et al. 2001; Dillin et al. 2002; Lee et al. 2003). For example, while the mutation of clk-1, a demethoxyubiquinone hydroxylase, which disrupts ubiquinone biosynthesis, extends lifespan, the mutation of mev-1, a cytochrome b subunit in complex II of mitochondria, enhances sensitivity to oxidative stresses and shortens lifespan (Ishii et al. 1990, 1998; Wong et al. 1995; Ewbank et al. 1997). It has been shown that autophagy functions to degrade mitochondria in mitochondrial homeostasis (Kim et al. 2007). It has also been reported that suppression of atp-3, a mitochondrial ATP synthase, or clk-1 cannot extend lifespan in the unc-51, bec-1 or atg-18 mutants, while RNAi of bec-1 after development has no effect on lifespan in long-lived clk-1 or isp-1, a Rieske iron sulfur protein of mitochondrial complex III, mutants (Hansen et al. 2008; Tóth et al. 2008). When we performed RNAi in a long-lived clk-1 mutant, RNAi of the three autophagy genes extended the lifespan of the clk-1(e2519) mutant markedly (Table 4). When we performed the same RNAi in a short-lived mev-1 mutant, RNAi of unc-51 or atg-9 extended the lifespan of the mev-1(kn1) mutant markedly (Table 4). So, mitochondrial function may not be involved in the autophagy gene suppression-induced lifespan extension.
Autophagy and apoptosis are linked together by several mechanisms (Kroemer & Levine 2008). We considered the possibility that suppression of autophagy genes causes the change of apoptosis and then affects lifespan. So we performed RNAi of autophagy genes in a caspase ced-3 mutant. RNAi of unc-51 or atg-9 extended the lifespan of the ced-3(n2433) mutant (Table 4), suggesting that caspase-mediated apoptosis may not be involved in the autophagy gene suppression-induced lifespan extension.
Discussion
In this study, we have shown that suppression of autophagy genes can extend lifespan in C. elegans. Our results show that although maternal suppression of seven autophagy genes in N2 reduces lifespan, maternal suppression of seven autophagy genes in daf-2 mutants extends lifespan and that suppression of each of at least three autophagy genes after development extends lifespan in both N2 and daf-2 mutants. We could suggest that suppression of autophagy genes can extend lifespan if it does not cause developmental abnormalities.
We also examined the effect of suppression of autophagy genes on lifespan in several mutants of the known pathways or genes that regulate lifespan in C. elegans. In these measurements, suppression of autophagy genes extended the lifespan of all the mutants examined, supporting our finding that suppression of autophagy genes can extend lifespan. Moreover, these results suggest that the lifespan extension by suppression of autophagy genes may not absolutely require the pathways or genes we analyzed.
It has been suggested that autophagy functions in cell death as well as in cell survival (Kroemer & Levine 2008). It has also been suggested that physiological levels of autophagy promote survival, whereas insufficient or excessive levels of autophagy promote death, for example, during starvation in C. elegans (Kang et al. 2007). Interestingly, it is also known that lifespan response depends on the levels of inhibition of genes encoding mitochondrial proteins in C. elegans (Rea et al. 2007); at low levels of inhibition, there is no response, and then as inhibition increases, lifespan begins to increase, but when inhibition becomes too severe, lifespan reduces. The change in the lifespan response to different levels of inhibition may also happen in the case of autophagy genes. So we may speculate that certain levels of inhibition of autophagy genes can extend lifespan while other levels of inhibition of autophagy genes can reduce lifespan or cannot affect lifespan. This could be one explanation for the difference in the lifespan changes after suppression of autophagy genes between our present study and previous studies.
Our results suggest that autophagy is not always beneficial to longevity and may have the function to restrict the lifespan of worms, although we cannot exclude the possibility that some of the autophagy genes would be involved in other functions than autophagy, which affect lifespan. It may be interesting to determine optimal levels of autophagy or autophagy genes for longevity.
Experimental procedures
Strains and growth condition
Caenorhabditis elegans strains were maintained at 20 °C on nematode growth medium (NGM) agar plates with Escherichia coli, OP50, as described (Brenner 1974). N2, daf-2(e1370), daf-16(mgDf50), sir-2.1(ok434), rsks-1(ok1255), mev-1(kn1), clk-1(e2519), ced-3(n2433) mutant strains and a sir-2.1 over-expression strain sir-2.1(geIn3) were sent by the Caenorhabditis Genetics Center. For a daf-16(mgDf50); sir-2.1(ok434) double mutant, sir-2.1(ok434) males were mated to daf-16(mgDf50) hermaphrodites. The following forward and reverse primers were used for PCR to investigate the presence of the deletion: for daf-16(mgDf50): 5′-CGA ATT CAG AAT GAA GGA GCC-3′ and 5′-GCT TGA AGT TAG TGC TTG GC-3′ (which generate a product of 1.1 kb in wild-type but not in the mutant); for sir-2.1(ok434): 5′-TTC AGA AGT TGC GGT CAC AC-3′ and 5′-GAT CAA ATG AGC ATT CGG CT-3′ (which generate a product of 2.3 kb in wild-type and a product of 1.5 kb in the mutant).
Search for autophagy genes in C. elegans
To identify autophagy genes in C. elegans, BLAST search in WormBase (http://www.wormbase.org/db/searches/blast_blat) was used. First, we searched for C. elegans autophagy genes by using yeast Saccharomyces cerevisiae autophagy genes which function in macroautophagy. When we could not find orthologs, we next searched for C. elegans autophagy genes by using mammalian autophagy genes, which are listed in the reference (Klionsky et al. 2003).
RNA interference
RNAi was performed essentially as described (Kamath et al. 2001). cDNA fragments of autophagy genes were amplified from C. elegans cDNA by PCR. The following forward and reverse primers were used: for unc-51: 5′-ATG TTG ATC GCA CAG ACG-3′ and 5′-TCT CGA GGA TTC TCT TCG-3′; for atg-3: 5′-CCG AGA GTC AAA ATT CCG-3′ and 5′-CAT AAG TTT GCT CGA CGG-3′; for atg-4.1: 5′-GGA AGA TGG AAT CGA GGC-3′ and 5′-ATT CCG ACG CAT TGT GGG-3′; for atg-4.2: 5′-CAG ATG ATG ATG GAC ACG-3′ and 5′-ATT GCA GCA ATT CCA ACC-3′; for atg-5: 5′-GTG GGA ATC GCA TGT TCC-3′ and 5′-GAC ATA AAC CCG TAG TGG-3′; for bec-1: 5′-TCT GAC TGG ACA TTC TCG-3′ and 5′-TGT TGT ACG CCG ATA CGC-3′; for atg-7: 5′-GAA ACT CGA TGA GAC GCC-3′ and 5′-ATC TGC TGA GCA TTC AGC-3′; for lgg-1: 5′-TGA AGT GGG CTT ACA AGG-3′ and 5′-CTT TCG TCA CTG TAG GCG-3′; for lgg-2: 5′-CAT TTC CGG AAT CGT TCC-3′ and 5′-CAA ATG CTG GTT GAG ACG-3′; for atg-9: 5′-TGA TGA TGA AGT GCT GCG-3′ and 5′-AGA TAG GCG ATT GGT CCG-3′; for atg-10: 5′-CAC AGA ACA ACA ATT CCG-3′ and 5′-TTT GTT GCC CAT ATA CGC-3′; for lgg-3: 5′-AGA AAC CGC CAC AAC TCC-3′ and 5′-TGA AGC TCC AGA ATC TCC-3′; for atg-16.2: 5′-GGC TGA CAG TGA ATC TCG-3′ and 5′-TGT ACG ATC CGC AGA TCC-3′; for atg-18: 5′-TCG ATC AAC TAC ATA GGC-3′ and 5′-CTG ATG AGA AGC ATA GCG-3′. Each fragment was cloned into the feeding vector pPD129.36. The RNAi constructs or empty vector (as control) were transformed into HT115 strain and the bacteria were seeded onto NGM plates containing isopropyl-β-D-thiogalactopyranoside (IPTG) and ampicillin.
Lifespan measurements
To synchronize worm populations, eggs were isolated by treating adults with a solution of sodium hypochlorite and NaOH. For maternal RNAi, the eggs were allowed to grow to L3–4 on normal NGM plates and transferred to RNAi plates for 2 days and the progeny were transferred to new RNAi plates. The progeny allowed to grow to young adult and transferred to new RNAi plates containing 5-fluoro-2′-deoxyuridine (FUDR) for lifespan measurements (about 20 worms per plate). For RNAi from young adult, i.e. after development, the eggs were allowed to grow to young adult on normal NGM plates and transferred to RNAi plates containing FUDR for lifespan measurements. Lifespan measurements were performed at 20 °C. Animals were scored every 2–3 days and counted as dead when they did not move after repeated taps with a pick. They were transferred to new RNAi plates every 4–5 days. Animals that crawled off the plate, displayed extruded internal organs or died from internally hatched progeny were excluded from the statistical analysis. We performed generally two plates in each trial and P-values were calculated by t-test using the lifespan data of each plate unless stated otherwise.
Acknowledgements
This work was supported by grants from Ministry of Education, Culture, Sports, Science and Technology of Japan (to E. N.). We thank the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources (NCRR), and the International C. elegans Gene Knockout Consortium for providing many C. elegans strains. We thank members of the Nishida laboratory for technical comments and helpful discussion.