Modeling human colon cancer in rodents using a food-borne carcinogen, PhIP
Abstract
Animal models provide researchers with powerful tools to elucidate multistage mechanisms for cancer development and to gain further insights into the biological roles of various cancer-related genes in in vivo situations. As for colon cancer models in rodents, Apc-disrupted mice, including ApcMin, have been one of the most widely utilized animal models to dissect the molecular events implicated in the development of intestinal tumors. In rats, several models have been established using chemical carcinogens, including azoxymethane and 2-amino-1-methyl-6-phenylimidazo- [4,5-b]pyridine (PhIP). The former is a representative colon carcinogenic alkylating agent, and the latter a heterocyclic amine produced while cooking meat and fish, which people are exposed to in ordinary life. It is of great importance to note that PhIP preferentially targets the colon and prostate gland in male rats, and the mammary glands in female rats. Cancers in these three organs are common in Western countries and are currently increasing in Japan, where modern dietary habits are rapidly becoming more like those of the West. In the present article, the history of PhIP-induced colon cancer models in rodents, activation/detoxification mechanisms of PhIP with regard to the formation of PhIP-DNA adducts, mechanistic approaches to dissect the molecular events involved in the development of colon cancer by PhIP, and epidemiological evidence of human exposure to PhIP are overviewed. The induction of Paneth cell maturation/differentiation in PhIP-induced colon cancers, genetic traits affecting susceptibility to colon carcinogenesis, and the biological relevance of colon cancer models in rodents to studying human colon carcinogenesis are also discussed. (Cancer Sci 2005; 96: 627 – 633)
Abbreviations:
-
- AB-PAS
-
- alcian blue-periodic acid Schiff
-
- ACF
-
- aberrant crypt foci
-
- BCAC
-
- β-catenin accumulated crypts
-
- HCA
-
- heterocyclic amine
-
- MDF
-
- mucin-depleted foci
-
- PhIP
-
- 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine
Human cancers are believed to be caused by the combined effects of both hereditary and environmental factors. Recently, Lichtenstein et al. published an important work that estimated the effects of genetic and environmental factors on common cancers using data from the Swedish, Danish and Finnish twin registries.(1)
Regarding hereditary factors, a considerable amount of research into familial cancers has revealed a series of tumor suppressor genes, including TP53, RB, APC, BRCA1/2, TSC1/2, WT1 and PTEN, comprising genetic traits with high-penetrance. However, we should also take low-penetrance genetic traits, for example genes in the CYP, NAT and GST families, into consideration. As for environmental factors, epidemiological studies have shown that smoking, infection and dietary habits play an important role in the development of sporadic human cancers.(2) Regarding human cancers related to infection and/or inflammation, examples are hepatitis B and C viruses in hepatocellular carcinoma, Helicobacter pylori in gastric cancer, and human papilloma virus in cervical cancer,(3) although molecular mechanisms underlying the onset of infection-related cancers still remain elusive. As for dietary factors, they are estimated to account for approximately one-third of human cancers.(2)
Concerning daily food intake, nitrites/nitrates may play some roles in relation to the formation of nitroso compounds, but the significance of nitroso compounds in human carcinogenesis is still unclear. Other important compounds in cooked foods are polycyclic aromatic hydrocarbons(4) and heterocyclic amine (HCA) compounds.(5,6) An example of the former is, for instance, benzo[a]pyren (B[a]P), which is present in roasted coffee beans.(4) HCA compounds are some of the most abundant genotoxic mutagens present in our environment, and humans are exposed to them in ordinary daily life. In this review article, the research history of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) (one of the most abundant HCA in heated meat and fish(7)) and a colon cancer model in rodents induced by PhIP are discussed from the viewpoint of their relevance to modeling human colon cancers.
PhIP, a food-borne carcinogen
Since the first discovery of HCA in 1977,(8) tremendous progress has since been made in this field in basic research. HCA in cooked food are produced by heating amino acids and proteins, and most HCA in food are really derived from heating meat. Heating meat yields various potent imidazoquinoline, imidazoquinoxaline and imidazopyridine compounds, and they are highly mutagenic towards some strains of Salmonella typhimurium. Some HCA have been synthesized in abundance, and have been found to be carcinogenic in long-term animal assays. PhIP was isolated by James Felton (Lawrence Livermore National Laboratory, California) by carefully examining a fraction with low peak mutagenicity.(7) Although the specific mutagenicity of PhIP is 100-fold less than that of IQ and MeIQ compounds, the PhIP content in cooked meat is much higher than that of the other two compounds.(9)
Metabolic activation and detoxification of PhIP
The structure of PhIP was determined with data from mass- and nuclear magnetic resonance spectrometries as depicted in Figure 1.(7) PhIP is soluble in dimethyl sulfoxide, stable under moderately acidic and alkaline conditions, and also under dry and cold conditions for a year. Once PhIP is given to animals, it is absorbed from the alimentary tract and is immediately and widely distributed throughout the body. PhIP is then metabolically activated by Phase I and Phase II enzymes mainly in the liver, similar to other HCA compounds, and probably in terminal target organs as well.(9,10) The principal pathway for metabolic activation of PhIP is shown in Figure 1. Cytochrome P450-mediated N-hydroxylation of PhIP occurs predominantly by CYP1A2 at liver microsomes.(9,10) N-Hydroxylated metabolites of PhIP are able to bind covalently to DNA to form adducts, but are further activated by Phase II enzymes, namely N- and O-acetyltransferases (NAT1 and NAT2) and sulfotransferases (SULT), to generate carcinogenic species. The Phase II enzymes add N-acetoxy or N-sulfonyl moieties to N-hydroxylated PhIP in the liver and partly in the terminal target organs, and arylnitrenium ions (R-NH+) derived from these ultimate metabolites of PhIP react with DNA to generate PhIP-adducted bases,(10) which results in the induction of mutations in the DNA replication process.
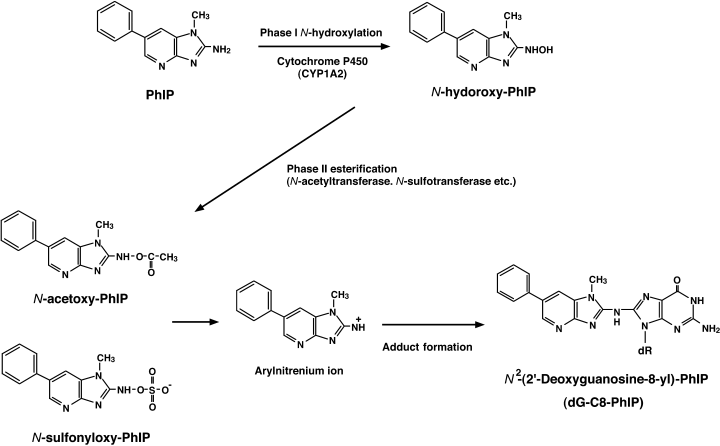
Chemical structure and pathway of metabolic activation of PhIP leading to DNA adduct formation. N-Hydroxylation of PhIP is catalyzed by phase I hepatic cytochrome P450, mainly by CYP1A2, followed by phase II esterification with N-acetyltransferase and N-sulfotransferase. Arylnitrenium ions generated from N-acetoxy-PhIP or N-sulfonyloxy-PhIP react with nucleophilic sites in DNA, mainly at guanine bases, leading to the formation of dG-C8-PhIP adducts.
As for a DNA-adduct of PhIP, the adduct in which the exocyclic amino group of PhIP covalently binds to the C8 atom of the guanine base (dG-C8-PhIP) is the major one found in calf thymus DNA reacted in vitro with N-acetoxy-PhIP,(9,10) and that found in DNA from rats exposed to PhIP. Regarding detoxification of PhIP, glutathione (GSH) and glutathione transferases (GST) play a key role. Furthermore, detoxification of N-hydroxylated PhIP occurs in the liver by generating glucuronate conjugates.(11) In this case, glucuronate-conjugated PhIP is secreted in the bile, deconjugated by gut flora, and reabsorbed from the gut (so-called ‘enterohepatic circulation’). This circulation makes colon epithelial cells receive repeated exposure to activated PhIP in vivo.
Evidence of human exposure to PhIP
The amounts of PhIP in cooked meat and fish range from 0.29 to 182 ng/g,(9) and the daily intake of PhIP is estimated to range between 0 and 865 ng/day, with a mean value of 72 ng/day.(12) To date, a considerable number of reports on the presence of low levels of both PhIP and PhIP-DNA adducts in human materials such as urine, feces and hair, have also been made. As for the levels of human exposure to PhIP, high performance liquid chromatography (HPLC) is able to directly and sensitively measure the PhIP in tissues at nanogram levels, and mass spectrometry (MS) gave a much higher sensitivity. Hashimoto et al. recently developed a column-switching liquid chromatography (LC)/MS method for quantification of PhIP in human hair samples, whose limit of quantification was 50 pg/g.(13) Using this method, PhIP at concentrations of 110–3878 pg/g was found in 42 of 46 (91.3%) hair samples from 23 healthy volunteers.(13)
Quantification of the levels of human exposure to PhIP can also be done by measuring the amount of PhIP-DNA adducts in tissue. A major DNA adduct of PhIP is dG-C8-PhIP, as described earlier, and 32P-postlabeling analysis,(14) HPLC(15) and an immunohistochemical method using an anti-PhIP-adducted DNA polyclonal antibody,(16) have been widely used for the detection of PhIP-adducted DNA. Gas chromatography (GC)/MS(17) and accelerator MS (AMS)(18) analyses have also been developed to detect a tiny amount of PhIP-DNA adducts, at sensitivities in the order of 10−8 and 10−12, respectively. PhIP-adducts detected in human colon samples by using the 32P-postlabeling method are approximately three adducts per 108 nucleotides.(17)
Mutagenic activity of PhIP
The mutagenic activity of HCA, which has been measured using a S. typhimurium histidine auxotroph, varies widely among various kinds of HCA molecules (Table 1). The mutagenicity of 2-amino-3-methyl-9H-pyrido[2,3-b]indole (MeAαC) in S. typhimurium TA98 that detects mutants with frameshift-type mutations gave as little as 200 revertants/µg compound. 2-Amino-3-methylimidazo[4,5-f]quinoline (MeIQ), in contrast, gave more than 6 × 105 revertants/µg. In S. typhimurium TA100, which detects mutants with base substitution-type mutations, the mutagenicity of HCA range from 20 revertants/µg for 2-amino-9H-pyrido[2,3-b]indole (AαC) to 3 × 104 revertants/µg for MeIQ. As for PhIP, it produced relatively low mutagenic activities relative to other HCA: 1800 in TA98 and 120 in TA100.(9) The mutagenicity of PhIP toward mammalian cells has also been evaluated using the Hprt or the Ef-2 genes as shuttle vectors. The mutant frequency of the Hprt gene in Chinese hamster fibroblast cells was approximately 90 × 10−6 with 100 µM PhIP, compared with 8 × 10−6 in the solvent control.(19)
| Compound | Revertants/µg | ||
|---|---|---|---|
| Abbreviation | Full name | TA98 | TA100 |
| IQ | 2-amino-3-methylimidazo[4,5-f]quinoline | 433 000 | 7000 |
| IQx | 2-amino-3,4-dimethylimidazo[4,5-f]quinoxaline | 75 000 | 1500 |
| MeIQ | 2-amino-3-methylimidazo[4,5-f]quinoline | 661 000 | 30 000 |
| MeIQx | 2-amino-3,4-dimethylimidazo[4,5-f]quinoxaline | 145 000 | 14 000 |
| 4,8-DiMeIQx | 2-amino-3,4,8-trimethylimidazo[4,5-f]quinoxaline | 183 000 | 8000 |
| PhIP | 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine | 1800 | 120 |
| AαC | 2-amino-9H-pyrido[2,3-b]indole | 300 | 20 |
| MeAαC | 2-amino-3-methyl-9H-pyrido[2,3-b]indole | 200 | 120 |
In vivo mutation spectra for PhIP have been measured in various organs using Big Blue rats(20,21) and mice,(22) gptΔ transgenic mice(23) and Muta mice,(24,25) and the values are summarized in Table 2. In the colon mucosa, a G-deletion is observed at a frequency of 26% and 34% in Big Blue mice and rats, respectively, and, especially, that from the 5′-GGGA-3′ sequence is somehow characteristic for PhIP. This accounts for 6.9% and 8.5% of the total mutations in the transgenic lacI gene in rats(20,21) and mice,(22) respectively, and 10% of all mutations detected in the Hprt gene in the Chinese hamster fibroblast as well.(19) As for base substitution-type mutations, G:C to T:A is the most common and accounts for approximately 25–50% of all mutations detected in the lacI gene of Big Blue animals.(20–22)
| Animal | Target gene | No. mutants (%) | ||||||
|---|---|---|---|---|---|---|---|---|
| One-base substitution | One-base deletion | Other types of mutations | Total | |||||
| G:C to T:A | G:C to others | Others | G:C | Others | ||||
| Big Blue rat | lacI* | 56 (27) | 57 (27) | 7 (3) | 72 (34)§ | 2 (1) | 16 (8) | 210 (100) |
| CII * | 61 (42) | 56 (39) | 2 (1) | 14 (10) | 3 (2) | 9 (6) | 145 (100) | |
| Big Blue mouse | lacI* | 56 (49) | 21 (18) | 4 (3) | 30 (26)¶ | 0 | 5 (4) | 115 (100) |
| Muta mouse | lacZ† | 13 (33) | 12 (30) | 1 (3) | 8 (20) | 1 (3) | 5 (13) | 40 (100) |
| CII ‡ | 23 (70) | 9 (27) | 0 | 1 (3) | 0 | 0 | 33 (100) | |
| gptΔ mouse | gpt * | 52 (53) | 27 (27) | 1 (1) | 13 (13) | 1 (1) | 5 (5) | 99 (100) |
| gam * | 10 (11) | 1 (1) | 0 | 73 (83) | 2 (2) | 2 (2) | 88 (100) | |
- * Large intestine;
- † † small and large intestine;
- ‡ ‡ small intestine;
- § § includes 18 cases (8.5%) of GGGA to GGA mutation;
- ¶ ¶ includes eight cases (6.9%) of GGGA to GGA mutation.
Target organs of PhIP
The in vivo carcinogenic potential of PhIP against rodents has been extensively investigated. Using a standard long-term experimental protocol, it preferentially targets the colon and prostate in male rats, mammary glands in female rats, and lymphoid tissues in both male and female rats.(26) It is important to note that human cancers in the colon, mammary glands and prostate gland are common in Western countries, and are currently increasing in Japan where dietary habits are becoming more Westernized. Regarding the tissue-specific carcinogenicity of PhIP in rats, Lauber et al. recently reported that steroid hormone receptors, such as estrogen receptor α (ERα), estrogen receptor β (ERβ) and progesterone receptor (PR), are upregulated in PhIP-induced tumors. PhIP has estrogenic action, stimulates cell proliferation in an ER-dependent manner, and activates the mitogen-activated protein kinase (MAPK) signaling pathway through mRNA induction of PR and c-myc genes.(27) This could be a causative genetic event conferring tumors with the sex hormone dependency of PhIP-induced tumors. Compared with rats, the organ spectrum of PhIP in mice is more prevalent and restricted to lymphoid tissues.(9)
Colon carcinogenesis in rodents by PhIP
As described earlier, the colon is one of the most preferred target organs for cancer in rats induced by PhIP, especially in male rats. In contrast, mice rarely develop colon cancers after standard long-term PhIP-feeding protocols. We previously reported the development of small-intestinal cancers in C57BL/6 N mice, but not colon cancers, with a short-term PhIP-feeding protocol with a high-fat diet. Molecular mechanisms underlying the topographic difference between rats and mice in terms of site preference for tumor development remain to be elucidated.
When rats were fed PhIP at concentrations of 25–400 p.p.m. in their diet, they developed aberrant crypt foci (ACF), candidate neoplastic lesions in the colon, shortly after exposure to PhIP. A subset of ACF features dysplastic components in cryptic cells, and these dysplastic ACF often demonstrate β-catenin accumulation in either the cytoplasm or nucleus. Some of them harbor mutations in the β-catenin gene at codons responsible for the stability of the β-catenin protein. In particular, lesions harboring β-catenin mutations and demonstrating nuclear accumulation of β-catenin tend to demonstrate more dysplastic features. We have categorized these lesions as ACF with high-grade dysplasia (‘high-grade dysplastic ACF’). Mutation spectra detected in dysplastic ACF, especially in high-grade dysplastic ones, are similar to those observed in colon cancers induced by PhIP, as detailed later. Furthermore, mucin detected by alcian blue-periodic acid Schiff (AB-PAS) staining is almost completely depleted in these high-grade dysplastic ACF.
As for the induction of colon cancers by long-term standard animal experiments, F344 male rats frequently develop colon carcinomas, the incidence being approximately 50% at 52 weeks when animals are fed continuously with a diet containing 400 p.p.m. PhIP.(26) With 100 p.p.m. PhIP in their diet, it takes approximately 2 years to get a ∼50% cancer incidence in the colon, but no colon carcinomas were observed in 2 years with 25 p.p.m. PhIP.(28) This result tells us that there is a clear dose-dependency in the induction of cancers in the colon. Colon cancers induced by PhIP occur preferentially in the middle or distal part of the colon, and feature mostly polypoid growth. Most of the tumors are histologically diagnosed as well-differentiated tubular adenocarcinomas, and the lesions rarely invade into submucosal layers or metastasize to other organs. Paneth cells are frequently observed in PhIP-induced colon cancers, as described later. Chronological multistep profiles for the development of colon cancers from normal colonic epithelial cells through ACF and dysplastic ACF deduced by sequential histological and genetic analyzes are shown in Figure 2.
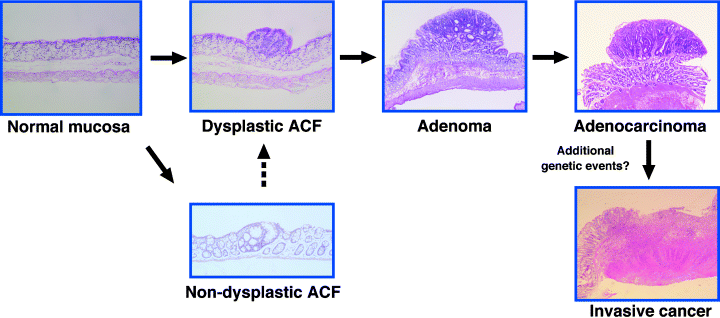
Multi-stage model of rat colon cancer induced by PhIP. ACF, a candidate preneoplastic lesion of the colon, are induced shortly after exposure to PhIP. The majority of ACF are histologically non-dysplastic lesions with a subset of lesions demonstrating dysplastic features. Approximately one-quarter of dysplastic ACF harbor either Apc or β-catenin mutations. Dysplastic ACF may further progress into more dysplastic lesions, namely high-grade dysplastic ACF, adenomas and adenocarcinomas. The susceptibility gene, Sct, on rat chromosome 16 is considered to control the development of ACF.
Modulation of PhIP-induced colon carcinogenesis
It is interesting to note that induction of colon cancers by a short-term intermittent feeding protocol of 400 p.p.m. PhIP in combination with a high-fat diet was almost equivalent to that observed by a conventional long-term PhIP (400 p.p.m.)-feeding protocol.(29) That is, 10- to 20-fold less PhIP was enough for colon cancer induction when the former protocol was adopted compared with the latter conventional protocol(29) (H Nakagama et al., unpublished observations). This indicates that experimental protocols themselves have a substantial effect on PhIP colon carcinogenesis. Furthermore, high-fat feeding accelerated the induction of colon carcinogenesis(30,31) as well as it did for mammary carcinogenesis induced by PhIP.(32) Diets high in calcium also showed a significant increase in the number of ACF and colon cancers compared with a low-calcium diet.(30) However, docosahexaenoic acid (DHA), a major component of fish oil, suppresses the formation of ACF and colon carcinomas after PhIP administration.(33)
Genetic alterations in PhIP-induced colonic lesions
Genetic analysis of PhIP-induced colon carcinomas has been carried out intensively by several groups, including ours.(34–36) Activation of the Wnt-β-catenin canonical signaling pathway is mainly caused by mutations in the β-catenin or Apc genes(34–39), similar to the process that occurs in human colon cancers.(40) Some of the lesions, however, do not have mutations in either of the genes.(29,39) Reduced expression of the Apc protein is also observed (H Nakagama et al., unpublished observations). Mutations in the β-catenin gene are commonly observed, mainly at codons 32, 34, 36 or 38 in exon 2 (exon 3 in the human β-catenin gene), the majority of which are G to T or G to A base substitutions (Table 3). Mutations in the Apc gene are somehow characteristic of PhIP, as has been described previously;(37) one G deletion occurs at G-stretch sequences (5′-GGGA-3′ to 5′-GGA-3′) in exons 14 and 15, and a G to T transversion mutation in a G stretch (5′-tagGGGGG-3′ to 5′-tatGGGGG-3′) occurs at the boundary of intron 10 and exon 11. This type of mutation at G-stretches is in good agreement with those observed as characteristic in vivo mutation spectra using shuttle vector systems.(41) In contrast to colon cancers induced in humans and in mice and rats by azoxymethane, which is another colon carcinogen widely used for cancer induction in animal experiments, K-ras mutations are rarely observed in PhIP-induced colon cancers.(42) Induction of p53 mutations is also a rare event in PhIP-induced colon cancers.(42,43)
| Gene | Mutation spectrum | No. lesions with mutations* | ||||
|---|---|---|---|---|---|---|
| Mutated position | Type of mutation | Amino acid change | Dysplastic ACF | High-grade dysplastic ACF | Colon cancer | |
| β-catenin | Codon 32 | GAT → AAT | Asp → Asn | 2 | ||
| GAT → GGT | Asp → Gly | 1 | 1† | |||
| Codon 34 | GGA → GTA | Gly → Val | 4 | 1 | 2 | |
| GGA → GAA | Gly → Glu | 3† | ||||
| Codon 36 | CAC → CCC | His → Pro | 1‡ | |||
| CAC → TAC | His → Tyr | 1 | ||||
| Codon 38 | GGT → CGT | Gly → Arg | 1‡ | |||
| Apc | Codon 1413in Exon 15 | GGGA → GGA | 1§ | |||
| Intron10/exon11 boundary | tagGGGGG→ tatGGGGG | 1 | 1 | |||
| Total no. lesions analyzed | 26 | 3 | 15 | |||
- * Colonic lesions are histologically classified into dysplastic ACF, high-grade dysplastic ACF (microadenoma) and colon cancers, and the number of lesions with specific mutations observed in each type of lesions are indicated;
- †‡ †‡ one case of colon cancer harbored mutations in both codons 32 and 34, and another one had them in both codons 36 and 38;
- § the same mutation was observed in colon cancers induced by long-term continuous feeding with PhIP.(37)
Another interesting observation is that Apc mutation frequency was significantly decreased from approximately 50% (4 of 8)(37) to 13% (2 of 15) by feeding rats with a high-fat diet with PhIP,(29,39) indicating that the promoting effect of a high-fat diet does not simply enhance the growth of cell populations with Apc mutations, but may also alter the repertoire of genetic alterations required for the development of PhIP colon cancers under a high-fat feeding regime. This phenomenon is reminiscent of the effect of dietary fat on Ha-ras gene mutations at codons 12 and 13 in PhIP-induced rat mammary gland tumors.(44)
Genomic instability induced by PhIP
PhIP has been reported to induce various types of genomic instability. In PhIP-induced colon cancers, microsatellite alterations were observed in some cases.(45) PhIP also induces size alterations of minisatellite DNA sequences,(46) which are composed of much longer repetitive units than microsatellite DNA and are dispersed in the genome. Intriguingly, Bardelli et al. reported that exposure to specific carcinogens could select tumor cells with distinct forms of genomic instability, and human colon cancer cells, HCT116 and DLD1, which became resistant to PhIP in culture conditions, exhibited a chromosomal instability.(47) Because PhIP produces a bulky adduct, in contrast to the alkylation of guanine bases by alkylating agents such as N-methyl-N1-nitro-N-nitrosoguanidine, PhIP-adducted DNA could result in the induction of chromosome breaks through nucleotide excision repair processes.(47) Later, Christian et al. demonstrated a cytogenetic signature of PhIP-induced mammary carcinomas using comparative genomic hybridization analysis,(48) but not in PhIP-induced colon cancers yet.
Gene expression profiles
Global gene expression analysis of PhIP-induced colon cancers using a high-density oligonucleotide microarray revealed that 27 and 46 of approximately 8800 genes or expressed sequence tags (EST) were over- and underexpressed, respectively, by threefold or greater in colon cancers.(49) For example, defensins, matrylysin (MMP7), macrophage metalloelastase (Mme) and cyclin D2 are highly expressed. In contrast, genes encoding carbonic anhydrase IV, mucin-like proteins, and muscle-related proteins are underexpressed, similar to the situation in human colon cancers. Insulin-like growth factor-binding protein gene (IGF-BP) and a gene related to supervillin were also markedly underexpressed.
A substantial number of genes in the list of over- and underexpressed genes in PhIP-induced colon cancers are either over- or underexpressed in human colon cancers as well (Table 4). For example, defensin family genes, α5 and α6, were previously reported to be upregulated in human colon cancers as noted in the SAGE database.(52) Furthermore, when six genes that are commonly overexpressed in PhIP colon cancers but have not been reported yet in human colon cancers, were analyzed by real-time reverse transcription–polymerase chain reaction (RT–PCR), four of them were demonstrated to be highly overexpressed in human colon cancers as well (H Nakagama et al., unpublished observations). Taking all these available data together, a substantial similarity does exist between colon cancers of humans and those induced in rats by PhIP.
| Gene name | Rat (fold change) | Human |
|---|---|---|
| Overexpressed | ||
| Defensin NP1 (α1)-like protein | 53.3 | Overexpressed† |
| Matrilysin (Mmp-7) mRNA | 41.9 | 8.0-fold |
| (EST) AA859937 | 14.4 | NA |
| Defensin NP3 (α3) gene | 13.5 | Overexpressed† |
| Platelet phospholipase A2 | 12.0 | Overexpressed |
| Type I keratin (Mhr a-1) | 9.9 | NA |
| Anti-acetylcholine receptor antibody gene | 9.6 | NA |
| Mash-2 mRNA expressed in neuronal precursor cells | 9.6 | Overexpressed |
| c-Ha-ras proto-oncogene mechanism sequence | 8.9 | NA |
| Intracellular calcium-binding protein Mrp14 | 8.2 | Overexpressed |
| Receptor-linked protein tyrosine phosphatase | 8.1 | NA |
| Macrophage metalloelastase (Mme) | 7.5 | 5.1-fold |
| Cation transporter Oct1A | 7.4 | NA |
| VL30 element | 6.8 | NA |
| Interleukin 1-β mRNA | 6.0 | Unchanged |
| Cyclin D2 | 5.6 | Overexpressed |
| α2-macroglobulin | 5.6 | Overexpressed |
| Underexpressed | ||
| (EST) AA799832 | 40.4 | NA |
| H36-α7 integrin α chain | 15.6 | 2.8-fold |
| Neuron-specific protein Pep-19 mRNA, complete cds | 13.6 | 2.1 |
| Sc1 protein | 12.2 | 4.5-fold |
| Zg-16p | 11.4 | NA |
| α, b-crystallin-related protein | 11.3 | NA |
| Dihydropyridine-sensitive L-type calcium channel | 10.4 | Unchanged |
| α-Crystallin B chain | 10.2 | 4.3-fold |
| (EST) AF119148/Filamin C, gamma | 9.6 | 5.8-fold |
| Alanine aminotransferase mRNA | 9.4 | NA |
| Insulin-like growth factor-binding protein | 9.3 | 1.6-fold |
| (EST) AA892888/ RIKEN cDNA 0610006F02 | 7.8 | NA |
| Carbonic anhydrase IV | 7.7 | 2-fold |
| (EST) AA800735/Supervillin (predicted) | 7.2 | NA |
| Guanylin | 6.0 | 20-fold |
| Phosphatase C-β1b | 5.8 | NA |
| Ssecks 322 | 5.7 | NA |
| D-binding protein | 5.1 | Underexpressed |
- Data for human cancers was drawn from various database and literature sources.(49–52)†Although α1 and α3 defensins have not been reported to be overexpressed in human colon cancer, α5 defensin, which is another member of the Paneth cell-specific defensins, is overexpressed in both adenomas and adenocarcinomas of the colon in humans.(51) NA, data not available.
Aberrant differentiation of Paneth cells in colon cancer
We also found that a subset of genes whose expression is characteristic of Paneth cells, namely the intestinal-type defensins and matrilysin, were overexpressed in colon cancers. Hematoxylin and eosin and AB-PAS staining revealed the presence of Paneth granules in colon cancer cells, and lysozyme expression was also observed in cells with Paneth granules.(49) Although molecular mechanisms underlying Paneth cell differentiation in colon cancer tissues have not been fully elucidated yet, activation of the Wnt/Apc/β-catenin signaling pathway could be a causative event. In fact, van Es et al. has recently reported activation of Wnt signaling to induce maturation of Paneth cells in intestinal crypts.(53) The appearance of Paneth cells may reflect the aberrant differentiation of colonic stem cells in cancer tissues.
Preneoplastic lesions of the colon
As for preneoplastic lesions of the colon, several candidates have been proposed. ACF, which were first reported by Bird in 1987 as precancerous lesions of the colon,(54) β-catenin accumulated crypts (BCAC)(55) and MDF(56) have been identified. Recently, flat dysplastic ACF was proposed as a strong candidate for preneoplastic lesions of the colon.(57) In the case of the classical type of ACF by Bird,(54) which appears shortly after PhIP administration, most of the lesions are non-dysplastic, without any mutations in the β-catenin or Apc genes.(39) A group of ACF harbor dysplastic components in cryptic cells, and are categorized as dysplastic ACF. A small fraction of dysplastic ACF that feature high-grade dysplasia lack mucin production in cells detected by AB-PAS staining. Lesional identities among high-grade dysplastic ACF, MDF, BCAC and flat dysplastic ACF are still a point of great and long dispute, but all of these lesions demonstrate β-catenin mutations and/or intense accumulation of β-catenin protein, and a drastic decrease in or depletion of mucin. Again, an appearance of Paneth cells is also occasionally observed in these dysplastic lesions, similar to the situation in colon cancers, as described earlier.(39)
Differential staining for dysplastic lesions in the colon
We have recently developed a novel and simple method to identify dysplastic ACF. By adding the step of a decolorization process with 70% methanol after conventional 0.2% methylene-blue staining, dysplastic ACF were easily and differentially contrasted as depicted in Figure 3A (‘Ochiai-Nakagama method’).(58) Furthermore, some of the dysplastic lesions became evident only after this differential staining procedure and, furthermore, the number of dysplastic ACF detected by this novel method more precisely reflected the carcinogenic potential of PhIP than the total number of ACF.(58) Examples are depicted in Figure 3Ba–f. Some of the lesions demonstrate heterogeneous features (Fig. 3Ba,b). The presence of this kind of lesion may suggest that the sequential transition from non-dysplastic to dysplastic ACF and also from dysplastic to high-grade dysplastic lesions within a lesion may occur in a subset of ACF. Lesions in Figures 3Bc–f demonstrate the typical features of high-grade dyplastic ACF. Both lesions characteristically demonstrated aberrant crypts with small or pin-holed orifices of crypts, and irregular alignments of crypts in the lesion. As for the lesion represented in Figure 3e, the lesion is very tiny when observed from the surface; however, histological examination revealed the actual size of this lesion to be much larger: it was embedded in the mucosal layer, with dysplastic components along with Paneth cells.
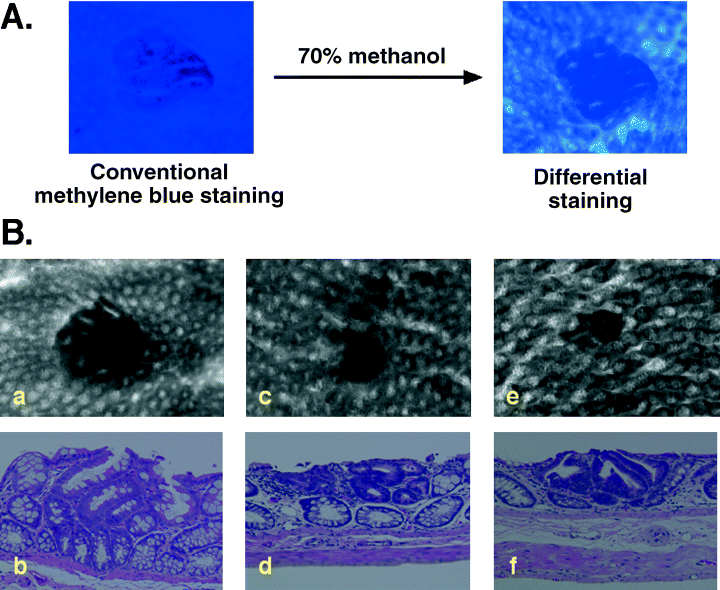
Differential staining, a simple and efficient method for selectively contrasting dysplastic lesions in the colon induced by PhIP. (A) For conventional methylene blue staining of ACF, strips of colon tissues are fixed in 10% neutralized formalin overnight at 4°C, and stained with 0.2% methylene blue in phosphate-buffered saline for 30 min. For differential staining, tissues strips are subsequently decolorized with 70% methanol with gentle shaking at room temperature for 4–6 min. Dysplastic ACF detected after the differential staining are characterized as lesions containing crypt(s) with homogeneously dense staining, and relatively small or even undetectable orifices of the crypts compared with normal crypts. Non-dysplastic ACF, in contrast, are composed of crypts with clear orifices and are stained relatively lightly compared to dysplastic ones. Some of the non-dysplastic lesions even became undetectable after the decolorization process. (B) Representative features of dysplastic ACF. (a,c,e) Digitized images of lesions detected by differential staining and (b,d,f) their respective histological features on hematoxylin and eosin staining are presented. Some dysplastic lesions (c,e), which show high-grade dysplasia (d,f), became evident only after the differential staining.
Differential susceptibility to colon carcinogenesis
Differential susceptibility to the development of colon cancers induced by PhIP is observed between male and female F344 rats,(25) although DNA-adduct levels and in vivo mutation frequencies measured using the lacI shuttle vector system are almost equivalent.(59) Ochiai et al. found that levels of cell proliferation in colon epithelial cells measured by BrdU incorporation after PhIP exposure were approximately 1.5-fold higher in male than in female rats, and this may confer a sexual difference.(59)
Strain differences in the susceptibility to PhIP colon carcinogenesis are also observed. The average number of ACF induced by PhIP varies widely among rat strains. BUF rats are highly sensitive, F344 and Brown-Norway are moderately sensitive, and ACI are resistant in terms of the induction of ACF.(60) F344 rats are indeed more susceptible to the induction of colon cancers compared with the ACI strain. Genetic traits responsible for differential susceptibility to the development of ACF by PhIP (we named this trait Sct[susceptibility to colon tumor]) were mapped on rat chromosome 16, between D16Rat17 and D16Wox3,(35) and research toward identification of the responsible gene(s) is currently being conducted intensively in our laboratory.
Epidemiological studies for human risk from PhIP
High-temperature cooking techniques and the ‘doneness’ level of red meat have been linked to the development of colorectal cancer, and Sinha and colleagues found that an elevated risk of colorectal adenomas is mainly due to an association with well-done/very well-done red meat consumption using an HCA database linked to a questionnaire to estimate HCA consumption.(61,62) Well-done meat has also been reported to be associated with increased risks of mammary and prostate gland carcinomas in humans.(9,63) Recently, Chao et al. further extended their study and reported that prolonged high levels of consumption of processed meat was associated with an increase in the risk of distal colon cancer after adjusting for age and energy intake.(64) Long-term consumption of poultry and fish was inversely associated with risk of both proximal and distal colon cancer, and high levels of consumption of red meat was associated with a higher risk of rectal cancer.(64) When considering human risk from PhIP, the effects of transplacental exposure to PhIP(65) and of PhIP-exposure in newborn babies on the generation of human cancers(66) should also be carefully investigated. In addition, the possible existence of a practical threshold value for the induction of cancers by PhIP(67) should be taken into consideration for assessment of its risk to humans, and further investigation is warranted in the future to clarify this point. It is practically important to note that because PhIP is the most abundant colon carcinogenic HCA in cooked meat and fish, to lessen the total amount of HCA intake from diet by, for example, avoiding high levels of consumption of red and processed meat and using a microwave oven or flipping meat during cooking, are recommended to decrease the chance of colon cancer development to some extent.
Future prospective views of the PhIP-induced colon cancer model
As is widely recognized, human cancers are caused by the combined effects of both hereditary and environmental factors. As for environmental factors, HCA are some of the most abundant genotoxic mutagens in our environment, and humans are exposed to them in their day-to-day lives. This has been evidenced by the presence of PhIP and PhIP-DNA adducts in human tissues. We therefore strongly believe that PhIP-induced colon cancer models in rats could serve as a powerful and relevant model system for the investigation of human colon carcinogenesis. Furthermore, numerous other compounds that can modulate the process of PhIP colon carcinogenesis, such as fat, inflammatory agents and other mutagenic/carcinogenic compounds, also exist abundantly in our environment. Although we do not have sufficient data on the combined effects of these environmental compounds on human carcinogenesis, synergistic effects from, for example, a mixture of various HCA and other non-genotoxic modulating compounds could reasonably be expected. This hypothesis could easily be recapitulated and evaluated in this rat model system. In fact, we have recently found that a combination of PhIP with some alkylating agents produced poorly differentiated and invasive adenocarcinomas in the rat colon. More detailed and comprehensive analysis should be conducted to assess the overall risk to humans from PhIP and also from other HCA in terms of its impact on human carcinogenesis. Moreover, Tanaka et al. have recently succeeded in inducing colon cancers using PhIP in mice in the presence of dextran sodium sulfate-induced colonic inflammation.(36) This mouse model is also of great benefit, and further extends the possibility and usefulness of PhIP-induced colon cancer models in rodents for the investigation of the molecular mechanisms of colon carcinogenesis, including those related to inflammation-related cancer development and those responsible for the genetic susceptibility of individual animals.
Acknowledgments
The authors are grateful to Drs Takashi Sugimura, Minako Nagao and Hiroshi Tazawa for critical reading of this article, and would also like to thank Kyoko Fujiwara, Ayako Taguchi and Ryoichi Masui for their continuing contribution to the work performed in our laboratory. This study was supported in part by Grants-in-Aid for Cancer Research and for the 2nd and 3rd Term of the Comprehensive 10-Year Strategy for Cancer Control from the Ministry of Health, Labour and Welfare of Japan, and by a Grant-in-Aid for Scientific Research on Priority Area (C) from the Ministry of Education, Culture, Sports, Science and Technology of Japan. MN was the recipient of a Research Resident Fellowship from the Foundation for Promotion of Cancer Research in Japan.




