Molecular phylogenetics of Paphiopedilum (Cypripedioideae; Orchidaceae) based on nuclear ribosomal ITS and plastid sequences
Abstract
Phylogenetic relationships in the genus Paphiopedilum were studied using nuclear ribosomal internal transcribed spacer (ITS) and plastid sequence data. The results confirm that the genus Paphiopedilum is monophyletic, and the division of the genus into three subgenera Parvisepalum, Brachypetalum and Paphiopedilum is well supported. Four sections of subgenus Paphiopedilum (Pardalopetalum, Cochlopetalum, Paphiopedilum and Barbata) are recovered as in a recent infrageneric treatment, with strong support. Section Coryopedilum is also recovered, with low bootstrap but high posterior probability values for support of monophyly. Relationships in section Barbata remain unresolved, and short branch lengths and the narrow geographical distribution of many species in the section suggest that it possibly underwent rapid radiation. Mapping chromosome and genome size data (including some new genome size measurements) onto the phylogenetic framework shows that there is no clear trend in increase in chromosome number in the genus. However, the diploid chromosome number of 2n = 26 in subgenera Parvisepalum and Brachypetalum suggests that this is the ancestral condition, and higher chromosome numbers in sections Cochlopetalum and Barbata suggest that centric fission has possibly occurred in parallel in these sections. The trend for genome size evolution is also unclear, although species in section Barbata have larger genome sizes than those in other sections. © 2012 The Linnean Society of London, Botanical Journal of the Linnean Society, 2012, 170, 176–196.
Introduction
The genus Paphiopedilum Pfitzer comprises c. 72 species (Averyanov et al., 2003), distributed from India and southern China through south-east Asia and the Malesian islands to the Solomon Islands (Cribb, 1998). Most species are terrestrial, but some are epiphytic or lithophytic (Cribb, 1998). This genus is the largest of the five genera of slipper orchids in subfamily Cypripedioideae (Orchidaceae). The other genera are Phragmipedium Rolfe, Selenipedium Rchb.f., Cypripedium L. and Mexipedium V.A.Albert & M.W.Chase. Floral characteristics of the slipper orchids are a slipper-shaped lip, two fertile stamens, a shield-like staminode and united lateral sepals or a synsepal (Cox et al., 1997). There is no unique morphological character to distinguish the slipper orchid genera from each other, but they can be distinguished by a combination of morphological characters, including leaf type, number of locules, type of placentation and geographical distribution (Cox et al., 1997). The characteristics of Paphiopedilum are conduplicate leaves, imbricate sepal aestivation and a unilocular ovary with parietal placentation. Paphiopedilum can be distinguished from the Northern Hemisphere Cypripedium and the tropical American Selenipedium by those genera having plicate leaves and perforate sepal aestivation. In addition, Selenipedium is distinguished by a trilocular ovary with axile placentation. Among the conduplicate leaved genera, Paphiopedilum can be distinguished from the central to southern American Phragmipedium by that genus having valvate sepal aestivation, a trilocular ovary and axile placentation and from the monotypic Mexipidium, which is restricted to Mexico, by Mexipedium having valvate sepal aestivation (Atwood, 1984; Albert & Chase, 1992; Cox et al., 1997).
The beautiful and often bizarre flowers of slipper orchids are not only attractive to insects but also to plant collectors, which have made them popular ornamental plants and has led to over-collection of plants from the wild, and this, along with the destruction of their habitat, means that many species are endangered or even facing extinction. The Convention on International Trade of Endangered Species (CITES) lists Paphiopedilum on Appendix I (CITES, 2012).
Paphiopedilum was first described by Pfitzer in 1886. Subsequently, infrageneric classifications of the genus have been proposed by various authors (Pfitzer, 1894, 1903; Hallier, 1896; Brieger, 1971; Karasawa & Saito, 1982; Atwood, 1984; Cribb, 1987, 1998; Braem, 1988; Cox et al., 1997; Braem, Baker & Baker, 1998; Averyanov et al., 2003; Braem & Chiron, 2003). An overview of previous infrageneric classifications is shown in Table 1.
| Pfitzer (1894) | Hallier (1896) | Pfitzer (1903) | Brieger (1971) | Karasawa & Saito (1982) | Atwood (1984) |
|---|---|---|---|---|---|
| Coelopedilum group | Coelopedilum group | ||||
| a. Eremantha Tessellata (in part) | Aphanoneura Brachypetalum | Brachypetalum | Brachypetalum | Brachypetalum | Brachypetalum |
| Parvisepalum | |||||
| b. Polyantha | Chromatoneura Viridia Polyantha | Anotopedilum | Polyantha | Polyantha | Paphiopedilum |
| XI Streptopetalum (in part) | Section Coryopedilum | Section Streptopetalum | Section Mastigopetalum | Section Coryopedilum | |
| XII Mastigopetalum | Section Gonatopedilum | Section Mastigopetalum | |||
| Section Prenipedilum | |||||
| Otopedilum | |||||
| XI Streptopetalum (in part) | Section Mystropetalum | Section Polyantha | Section Mystropetalum | Section Pardalopetalum | |
| X Pardalopetalum | Section Pardalopetalum | Section Polyantha | |||
| XIII Cochlopetalum | Section Cochlopetalum | Section Cochlopetalum | Cochlopetalum | Section Cochlopetalum | |
| a. Eremantha Viridia | Chromatoneura Viridia Eremantha | Paphiopedilum | Paphiopedilum | Section Paphiopedilum | |
| VIII Stictopetalum | Section Stictopetalum | Section Stictopetalum | Section Stictopetalum | ||
| IX Neuropetalum | Section Neuropetalum | Section Paphiopedilum | Section Paphiopedilum | ||
| V Thiopetalum | Section Thiopetalum | Section Thiopetalum | |||
| VII Cymatopetalum | Section Cymatopetalum | ||||
| VI Ceratopetalum | Section Ceratopetalum | Section Ceratopetalum | |||
| a. Eremantha Tessellata (in part) | Chromatoneura Tessellata | Barbata | Sigmatopetalum | Section Barbata | |
| II Sigmatopetalum | Section Spathopetalum | Section Sigmatopetalum | Section Spathopetalum | ||
| IV Drepanopetalum | Section Blepharopetalum | Section Blepharopetalum | Section Sigmatopetalum | ||
| Section Blepharopetalum | |||||
| Section Punctatum | |||||
| Section Planipetalum | |||||
| III Clinopetalum | Section Phacopetalum | Section Barbata | Section Barbata |
| Cribb (1987) | Braem (1988), Braem et al. (1998) and Braem & Chiron (2003) | Cox et al. (1997) | Cribb (1998) | Averyanov et al. (2003) |
|---|---|---|---|---|
| Brachypetalum | Brachypetalum | Brachypetalum | Brachypetalum | Brachypetalum |
| Section Brachypetalum | ||||
| Section Parvisepalum | Parvisepalum | Parvisepalum | Parvisepalum | Parvisepalum |
| Section Parvisepalum | ||||
| Section Emersonianum | ||||
| Paphiopedilum | Polyantha | Paphiopedilum | Paphiopedilum | Paphiopedilum |
| Section Coryopedilum | Section Mastigopetalum | Section Coryopedilum | Section Coryopetalum | |
| Section Pardalopetalum | Section Mystropetalum | Section Pardalopetalum | Section Pardalopetalum | Section Pardalopetalum |
| Section Polyantha | ||||
| Section Cochlopetalum | Cochlopetalum | Section Cochlopetalum | Section Cochlopetalum | Section Cochlopetalum |
| Section Paphiopedilum | Paphiopedilum | Section Paphiopedilum | Section Paphiopedilum | Section Paphiopedilum |
| Section Stictopetalum | ||||
| Section Paphiopedilum | ||||
| Section Thiopetalum | ||||
| Section Ceratopetalum | ||||
| Section Barbata | Sigmatopetalum | Section Barbata | Section Barbata | Section Barbata |
| Section Spathopetalum | ||||
| Section Sigmatopetalum | ||||
| Section Blepharopetalum | ||||
| Section Punctatum | ||||
| Section Planipetalum | ||||
| Section Barbata |
The first comprehensive study of the molecular phylogenetics of subfamily Cypripedioideae was that of Cox et al. (1997), using nuclear ribosomal DNA internal transcribed spacer (ITS) sequence data. The circumscriptions of sections in Paphiopedilum were, in general, congruent with the previous infrageneric classification of Cribb (1987). However, the result did not support the division of the genus into two subgenera, Brachypetalum (Hallier f.) Pfitzer and Paphiopedilum K.Karas. & K.Saito, because subgenus Brachypetalum was found to be paraphyletic to subgenus Paphiopedilum. Section Concoloria (Kraenzl.) V.A.Albert & Börge Pett. (=section Brachypetalum sensu Cribb, 1987) of subgenus Brachypetalum was nested in a clade of subgenus Paphiopedilum. In addition, section Coryopedilum Pfitzer was weakly supported as paraphyletic to the monophyletic section, Pardalopetalum Hallier f. & Pfitzer. Cox et al. (1997) tentatively proposed elevating section Parvisepalum (K.Karas. & K.Saito) P.J.Cribb and section Concoloria of subgenus Brachypetalum to subgenera Parvisepalum K.Karas. & K.Saito and Brachypetalum, and suggested combining sections Coryopedilum and Pardalopetalum in their infrageneric treatment. Also, they suggested simplification of the subsectional treatment of Braem (1988), because groupings of only a few species are less useful in understanding the relationships among the groups. Although the ITS results of Cox et al. (1997) suggested that the infrageneric classification of Cribb (1987) was mainly well defined, it did not provide support for monophyly of the largest subgenus, Paphiopedilum. In addition, the phylogenetic relationships between sections in subgenus Paphiopedilum remained unclear, because the resulting tree did not have sufficient bootstrap support for those clades.
The infrageneric classification of Cribb (1998) in his second edition of the monograph, mainly based on morphological characters and chromosome data, also followed the molecular study of Cox et al. (1997). Cribb subdivided Paphiopedilum into three subgenera in his classification: Parvisepalum; Brachypetalum; and Paphiopedilum. Five sections of subgenus Paphiopedilum (Coryopedilum, Pardalopetalum, Cochlopetalum Hallier f. ex Pfitzer, Paphiopedilum and Barbata (Kraenzl.) V.A.Albert & Börge Pett.) remained, as in his previous treatment.
Averyanov et al. (2003) followed the outline of the infrageneric classification of Cribb (1998), but they further divided subgenus Parvisepalum into two sections: Parvisepalum and Emersonianum Aver. & P.J.Cribb. The new section Emersonianum was recognized to include P. hangianum Perner & O.Gruss and P. emersonii Koop. & P.J.Cribb, which were differentiated mainly by these species having plain green leaves, whereas species of section Parvisepalum have tessellated leaves.
The analysis of nuclear DNA regions alone, such as ITS, as in the study of Cox et al. (1997), may be inadequate for obtaining the necessary resolution of phylogenetic relationships at lower levels, although they may evolve rapidly (e.g. Álvarez & Wendel, 2003). Sequence data from other loci, such as plastid DNA, can be useful for investigating the relationships between closely related species. Although generally evolving relatively slowly, various regions of the plastid genome have undergone more rapid evolution, potentially providing more variation for studying closely related taxa (e.g. Shaw et al., 2005, 2007). These data can also be utilized to test phylogenetic relationships independently and can be combined with data from other loci. Furthermore, unlike nuclear loci, plastid loci are uniparentally inherited (maternally in the case of slipper orchids, as for most flowering plants; Corriveau & Coleman, 1988), thus avoiding the potential problem of paralogous copies found in the nuclear genome.
In a recently published paper (Guo et al., 2012), six plastid DNA regions and two low-copy nuclear genes were used to study phylogenetics and biogeography in subfamily Cypripedioideae. As in earlier studies, Paphiopedilum was shown to be monophyletic, and it was strongly supported as sister to Phragmipedium/Mexipedium. Sampling of Paphiopedilum spp., however, was rather sparse (eight species only) and the focus was on relationships between, rather than within, the genera.
Genome size in angiosperms varies c. 2400-fold, from that of the carnivorous plant Genlisea margaretae Hutch. (Lentibulariaceae), 1C-value of only 0.0648 pg, to that of the monocot Paris japonica (Franch. & Sav.) Franch. (Melanthiaceae), the largest known genome of 1C = 152.23 pg (Greilhuber et al., 2006; Pellicer, Fay & Leitch, 2010; Bennett & Leitch, 2011). Most angiosperms have a small genome size; based on an analysis of > 6000 species, the modal and median of 1C values are only 0.6 and 2.9 pg (Bennett & Leitch, 2010). Species with very large genome sizes (i.e. 1C ≥ 35 pg, Kelly & Leitch, 2011) are found mainly in monocots, including Orchidaceae. Among angiosperms, based on available data, Orchidaceae have the greatest variation in genome size, ranging 168-fold from 1C = 0.33 pg in Oncidium maduroi Dressler to 55.4 pg in Pogonia ophioglossoides (L.) Ker Gawl. (Leitch et al., 2009).
Many species of subfamily Cypripedioideae have large genome sizes ranging > 10-fold from 1C = 4.1 pg in Cypripedium molle Lindl. to 43.1 pg in C. fargesii Franch., and Cypripedium is the most variable genus in the subfamily (Kahandawala, 2009; Leitch et al., 2009). Paphiopedilum spp. also have large genome sizes, ranging nearly two-fold, from 1C = 17.80 pg in P. godefroyae (God.-Leb.) Stein to 34.53 pg in P. wardii Summerh., whereas Phragmipedium spp. have smaller genomes and a narrower range, varying 1.5-fold, from 1C = 6.1 to 9.18 pg (Cox et al., 1998).
A considerable amount of chromosome data is available for Paphiopedilum (e.g. Karasawa, 1978, 1979, 1982, 1986; Karasawa & Aoyama, 1980, 1988; Karasawa & Tanaka, 1980, 1981; Karasawa & Saito, 1982; Karasawa, Aoyama & Kamimura, 1997; Cox et al., 1998). The diploid chromosome number in the genus varies from 2n = 26 to 42. All species so far analysed in the subgenera Parvisepalum and Brachypetalum have a chromosome number of 2n = 26, and many species in subgenus Paphiopedilum also have 2n = 26. In section Paphiopedilum, most species have 2n = 26, except for two species, which have 2n = 30. Chromosome numbers in section Cochlopetalum range from 30 to 37, and section Barbata is the most variable, with chromosome numbers ranging from 2n = 28 to 42. Despite the variation in chromosome number, the total number of chromosome arms (‘nombre fondamental’ or n.f., Matthey, 1949) appears to be conserved in most species of the genus (n.f. = 52), which might suggest karyotype evolution via Robertsonian change, either producing telocentric chromosomes by centric fission or producing metacentric chromosomes by centric fusion (Robertson, 1916). The first report to postulate Robertsonian change as a cause of total arm number retention in Paphiopedilum was that of Duncan & MacLeod (1949). Cox et al. (1998) studied the evolution of genome size and karyotype in Cypripedioideae by mapping chromosome number and genome size data onto a phylogenetic tree based on ITS data (Cox et al., 1997). The results for Paphiopedilum showed evolutionary trends of an increase in the number of chromosomes and telocentric chromosomes and a decrease in metacentric chromosomes, suggesting the predominant direction of karyotype evolution was via centric fission, leading to higher chromosome numbers. It also showed an increase in genome size. However, the phylogenetic tree used for their study did not provide support for phylogenetic relationships between sections of Paphiopedilum, as mentioned previously, and these hypotheses need to be reassessed in a phylogenetic framework with better resolution and support.
The aims of this study were to collect DNA sequence data from nuclear (ITS) and plastid (partial matK, ycf1, psaA-ycf3ex3 and trnF(GAA)-ndhJ) loci to address generic, subgeneric and sectional circumscription and to investigate phylogenetic relationships within the genus. In addition, the more robust phylogenetic trees were used as a framework to analyse evolutionary trends in genome size and chromosome number in the genus.
Material and Methods
Plant material
Most DNA samples were obtained from the DNA Bank at the Jodrell Laboratory (RBG, Kew). In addition, some leaf material was obtained for DNA extraction from the living plant collection at the Tropical Nursery, (RBG, Kew). As samples for two species, P. hangianum and P. emersonii, of subgenus Parvisepalum section Emersonianum in the treatment of Averyanov et al. (2003), were not available, we were not able to address the question on the monophyly of this group. The taxon sampling used in this study was based on the infrageneric treatment of Cribb (1998) for sampling subgenera Parvisepalum, Brachypetalum and Paphiopedilum (sections Coryopedilum, Pardalopetalum, Cochlopetalum, Paphiopedilum and Barbata). The morphological terms used also follow Cribb (1998). Outgroup taxa were sampled from Phragmipedium, the sister genus of Paphiopedilum (Cox et al., 1997). All species of Paphiopedilum and the outgroups used in this study, with voucher information, are listed in Table 2.
| Taxa | Voucher/source | GenBank accession numbers | ||||
|---|---|---|---|---|---|---|
| ITS | matK | ycf1 | psaA-ycf3ex3 | trnF(GAA)-ndhJ | ||
| Subgenus Parvisepalum | ||||||
| Paphiopedilum delenatii Guillaumin | Chochai 39746 (K) | JQ929314 | JQ929368 | JQ929521 | JQ929419 | JQ929470 |
| Paphiopedilum malipoense S.C.Chen & Z.H.Tsi | Z6 | JQ929336 | JQ929388 | JQ929541 | JQ929439 | JQ929490 |
| Paphiopedilum micranthum Tang & F.T.Wang | M.W. Chase O-629 (K) | JQ929338 | JQ929390 | JQ929543 | JQ929441 | JQ929492 |
| Subgenus Brachypetalum | ||||||
| Paphiopedilum concolor (Bateman) Pfitzer (a) | Z17 | JQ929312 | JQ929367 | JQ929520 | JQ929418 | JQ929469 |
| Paphiopedilum concolor (Bateman) Pfitzer (b) | Yang Ping, Guizhem. Luo s.n. | JQ929313 | – | – | – | – |
| Paphiopedilum niveum (Rchb.f.) Stein | 36862a, Kew 1990–996b (no voucher) | JQ929339 | JQ929391 | JQ929544 | JQ929442 | JQ929493 |
| Subgenus Paphiopedilum | ||||||
| Section Paphiopedilum | ||||||
| Paphiopedilum hirsutissimum (Lindl. ex Hook.) Stein | Chochai 36808 (K) | JQ929327 | – | – | – | – |
| Paphiopedilum hirsutissimum (Lindl. ex Hook.) Stein var. esquirolei (Schltr.) K.Karas. & K.Saito | M.W. Chase O-642 (K) | JQ929328 | – | – | – | – |
| Paphiopedilum charlesworthii (Rolfe) Pfitzer | M.W. Chase O-632 (K) | JQ929310 | JQ929365 | JQ929518 | JQ929416 | JQ929467 |
| Paphiopedilum insigne (Wall. ex Lindl.) Pfitzer | Chochai 36821 (K) | JQ929329 | JQ929381 | JQ929534 | JQ929432 | JQ929483 |
| Paphiopedilum exul (Ridl.) Rolfe | 36804a, Kew 1977–2853b (no voucher) | JQ929317 | JQ929371 | JQ929524 | JQ929422 | JQ929473 |
| Paphiopedilum gratrixianum (Mast.) Rolfe (a) | Chochai 36809 (K) | JQ929322 | JQ929376 | JQ929529 | JQ929427 | JQ929478 |
| Paphiopedilum gratrixianum (Mast.) Rolfe (b) | Chochai 40235 (K) | JQ929323 | JQ929377 | JQ929530 | JQ929428 | JQ929479 |
| Paphiopedilum gratrixianum (Mast.) Rolfe (c) | Chochai 40236 (K) | JQ929324 | JQ929378 | JQ929531 | JQ929429 | JQ929480 |
| Paphiopedilum villosum (Lindl.) Stein var. boxallii (Rchb.f.) Pfitzer | Chochai 36822 (K) | JQ929354 | JQ929405 | JQ929558 | JQ929456 | JQ929507 |
| Paphiopedilum tigrinum Koop. & N.Haseg. | ex Paul Phillips-Rathcliffe | JQ929351 | – | – | – | – |
| Paphiopedilum druryi (Bedd.) Stein | Chochai 36811 (K) | JQ929316 | JQ929370 | JQ929523 | JQ929421 | JQ929472 |
| Paphiopedilum spicerianum (Rchb.f.) Pfitzer | M.W. Chase O-643 (K) | JQ929347 | JQ929399 | JQ929552 | JQ929450 | JQ929501 |
| Section Barbata | ||||||
| Paphiopedilum appletonianum (Gower) Rolfe | M.W. Chase 5897 (K) | JQ929306 | JQ929362 | JQ929515 | JQ929413 | JQ929464 |
| Paphiopedilum sangii Braem | O-822a (no voucher) | JQ929346 | JQ929398 | JQ929551 | JQ929449 | JQ929500 |
| Paphiopedilum mastersianum (Rchb.f.) Stein | M.W. Chase 5900 (K) | JQ929337 | JQ929389 | JQ929542 | JQ929440 | JQ929491 |
| Paphiopedilum violascens Schltr. | O-825a (no voucher) | JQ929355 | JQ929406 | JQ929559 | JQ929457 | JQ929508 |
| Paphiopedilum tonsum (Rchb.f.) Stein | M.W. Chase 5902 (K) | JQ929352 | JQ929403 | JQ929556 | JQ929454 | JQ929505 |
| Paphiopedilum barbatum (Lindl.) Pfitzer | M.W. Chase 5898 (K) | JQ929307 | JQ929363 | JQ929516 | JQ929414 | JQ929465 |
| Paphiopedilum callosum (Rchb.f.) Stein | Z4 | JQ929308 | JQ929364 | JQ929517 | JQ929415 | JQ929466 |
| Paphiopedilum callosum (Rchb.f.) Stein var. sublaeve (Rchb.f.) P.J.Cribb | Z32 | JQ929309 | – | – | – | – |
| Paphiopedilum hennisianum (M.W.Wood) Fowlie | Z30 | JQ929326 | JQ929380 | JQ929533 | JQ929431 | JQ929482 |
| Paphiopedilum fowliei Birk | M.W. Chase O-644 (K) | JQ929318 | JQ929372 | JQ929525 | JQ929423 | JQ929474 |
| Paphiopedilum javanicum (Reinw. ex Lindl.) Pfitzer var. virens (Rchb.f) Stein | M.W. Chase O-635 (K) | JQ929330 | JQ929382 | JQ929535 | JQ929433 | JQ929484 |
| Paphiopedilum lawrenceanum (Rchb.f.) Pfitzer | Chochai 36824 (K) | JQ929332 | JQ929384 | JQ929537 | JQ929435 | JQ929486 |
| Paphiopedilum ciliolare (Rchb.f.) Stein | Z25 | JQ929311 | JQ929366 | JQ929519 | JQ929417 | JQ929468 |
| Paphiopedilum superbiens (Rchb.f.) Stein var. curtisii Braem | Z5 | JQ929350 | JQ929402 | JQ929555 | JQ929453 | JQ929504 |
| Paphiopedilum sukhakulii Schoser & Senghas | M.W. Chase 5901 (K) | JQ929349 | JQ929401 | JQ929554 | JQ929452 | JQ929503 |
| Paphiopedilum wardii Summerh. | M.W. Chase 5903 (K) | JQ929356 | JQ929407 | JQ929560 | JQ929458 | JQ929509 |
| Section Pardalopetalum | ||||||
| Paphiopedilum dianthum Tang & F.T.Wang | Z23 | JQ929315 | JQ929369 | JQ929522 | JQ929420 | JQ929471 |
| Paphiopedilum parishii (Rchb.f.) Stein | Z3 | JQ929340 | JQ929392 | JQ929545 | JQ929443 | JQ929494 |
| Paphiopedilum lowii (Lindl.) Stein (a) | Z22 | JQ929334 | JQ929386 | JQ929539 | JQ929437 | JQ929488 |
| Paphiopedilum lowii (Lindl.) Stein (b) | Chochai 36810 (K) | JQ929335 | JQ929387 | JQ929540 | JQ929438 | JQ929489 |
| Paphiopedilum haynaldianum (Rchb.f.) Stein | M.W. Chase O-175 (K) | JQ929325 | JQ929379 | JQ929532 | JQ929430 | JQ929481 |
| Section Cochlopetalum | ||||||
| Paphiopedilum glaucophyllum J.J.Sm. | Z21 | JQ929321 | JQ929375 | JQ929528 | JQ929426 | JQ929477 |
| Paphiopedilum liemianum (Fowlie) K.Karas. & K.Saito | 36858a, Kew 1990–8000b (no voucher) | JQ929333 | JQ929385 | JQ929538 | JQ929436 | JQ929487 |
| Paphiopedilum primulinum M.W.Wood & P.Taylor | Chochai 36827 (K) | JQ929342 | JQ929394 | JQ929547 | JQ929445 | JQ929496 |
| Paphiopedilum primulinum M.W.Wood & P.Taylor var. purpurascens (M.W.Wood) P.J.Cribb | 36860a, Kew 2001–3172b (no voucher) | JQ929343 | JQ929395 | JQ929548 | JQ929446 | JQ929497 |
| Paphiopedilum victoria-regina (Sander) M.W.Wood | M.W. Chase O-630 (K) | JQ929353 | JQ929404 | JQ929557 | JQ929455 | JQ929506 |
| Section Coryopedilum | ||||||
| Paphiopedilum philippinense (Rchb.f.) Stein | Chochai 36807 (K) | JQ929341 | JQ929393 | JQ929546 | JQ929444 | JQ929495 |
| Paphiopedilum randsii Fowlie | M.W. Chase O-636 (K) | JQ929344 | JQ929396 | JQ929549 | JQ929447 | JQ929498 |
| Paphiopedilum kolopakingii Fowlie | Z18 | JQ929331 | JQ929383 | JQ929536 | JQ929434 | JQ929485 |
| Paphiopedilum stonei (Hook.) Stein | Z7 | JQ929348 | JQ929400 | JQ929553 | JQ929451 | JQ929502 |
| Paphiopedilum adductum Asher | 36820a, Kew 1992–3661b (no voucher) | JQ929305 | JQ929361 | JQ929514 | JQ929412 | JQ929463 |
| Paphiopedilum glanduliferum (Blume) Stein (a) | M.W. Chase O-716 (K) | JQ929319 | JQ929373 | JQ929526 | JQ929424 | JQ929475 |
| Paphiopedilum glanduliferum (Blume) Stein (b) | M.W. Chase O-717 (K) | JQ929320 | JQ929374 | JQ929527 | JQ929425 | JQ929476 |
| Paphiopedilum wilhelminiae L.O.Williams | 36825a, Kew 2005–2702b (no voucher) | JQ929357 | JQ929408 | JQ929561 | JQ929459 | JQ929510 |
| Paphiopedilum rothschildianum (Rchb.f.) Stein | Chochai 36806 (K) | JQ929345 | JQ929397 | JQ929550 | JQ929448 | JQ929499 |
| Outgroup | ||||||
| Phragmipedium besseae Dodson & J.Kuhn | Z16a | JQ929358 | JQ929409 | JQ929562 | JQ929460 | JQ929511 |
| Phragmipedium schlimii (Linden ex Rchb.f.) Rolfe | M.W. Chase O-183 (VA) | JQ929360 | JQ929411 | JQ929564 | JQ929462 | JQ929513 |
| Phragmipedium longifolium (Warsz. & Rchb.f.) Rolfe | Z9 | JQ929359 | JQ929410 | JQ929563 | JQ929461 | JQ929512 |
- a Kew DNA bank number.
- b Kew living collection number.
Molecular Study
DNA extraction
For additional DNA samples, genomic DNA was extracted from fresh plant material, following the modified 2 × cetyl trimethylammonium bromide (CTAB) method of Doyle & Doyle (1987). DNA samples were purified by either caesium chloride/ethidium bromide density gradients or DNA purification columns (NucleoSpin Extract II Columns; Macherey-Nagel, GmbH & Co. KG, Germany) according to the manufacturer's protocols.
Amplification
The nuclear ribosomal spacers, ITS1 and ITS2, and the 5.8S ribosomal gene were amplified using the primers of Sun et al. (1994) and White et al. (1990). Partial matK, approximately 800 bp in length, was amplified using the primers of Sun, McLewin & Fay (2001). An approximately 1500-bp portion from the 3′ end of ycf1 was amplified using the primers of Neubig et al. (2009). The non-coding plastid regions, psaA-ycf3ex3 and trnF(GAA)-ndhJ, were amplified using the primers of Ebert & Peakall (2009).
All amplified PCR samples were purified using NucleoSpin Extract II columns according to the manufacturer's protocols. The PCR product was then sequenced using a Big Dye Terminator kit (Applied Biosystems Inc., Warrington, UK). The cycle sequencing products were cleaned by ethanol precipitation and then run on an ABI 3730 automated sequencer. Raw sequences were edited and assembled using Sequencher 4.1 software (Gene Codes Inc., Ann Arbor, MI, USA). The resulting sequences were then aligned manually. All sequences were deposited in GenBank.
Parsimony analysis
Sequence data were analysed independently and in combination, using the maximum parsimony criterion in PAUP* version 4.0b10 for Macintosh (Swofford, 2002). All characters were treated as unordered and equally weighted (Fitch, 1971). Parsimony analyses were conducted using a heuristic search strategy, with 1000 replicates of random taxon addition, tree–bisection–reconnection (TBR) branch swapping with MulTrees in effect, gaps treated as missing data and saving no more than ten trees per replicate. Support for groups was evaluated using 1000 replicates of bootstrap (Felsenstein, 1985), with simple addition and TBR swapping, saving ten trees per replicate. Groups were retained when bootstrap percentages (BP) ≥ 50.
Bayesian analysis
The best-fit models for nucleotide substitution for the data matrix of each region were determined by the Akaike information criterion test (Akaike, 1974) as implemented in MrModeltest version 2.2 (Nylander, 2004). The general time reversible model of substitution with gamma distribution (GTR + G) was selected for ITS, partial matK and psaA-ycf3ex3 data and the general time reversible model of substitution with gamma distribution and invariable sites (GTR + I + G) was selected for ycf1 and trnF(GAA)-ndhJ data.
All analyses were carried out using the parallel version of MrBayes version 3.1.2 (Ronquist & Huelsenbeck, 2003) through the University of Oslo Bioportal (http://www.bioportal.uio.no). Two runs of four Monte Carlo Markov chains (MCMC; Yang & Rannala, 1997) were performed for 10 000 000 generations and a tree was sampled every 1000 generations. Each parameter estimation obtained from the results of two runs was checked in Tracer version 1.5 (http://tree.bio.ed.ac.uk/software/tracer) to ascertain whether they had obtained proper effective sample size and to verify that stationary state had been reached. Trees from the first 10% of generations were discarded as burn-in. The remaining trees were combined to build a 50% majority-rule consensus tree in PAUP* version 4.0b10.
Chromosome number and genome size data
Chromosome numbers for Paphiopedilum and Phragmipedium were taken from the literature (Karasawa, 1979, 1980, 1982, 1986; Karasawa & Aoyama, 1980, 1988; Karasawa et al., 1997; Cox et al., 1998; Bennett & Leitch, 2010; Lan & Albert, 2011). Most genome size data were taken from the literature (Narayan, Parida & Vij, 1989; Cox et al., 1998; Bennett & Leitch, 2010). Seven species were measured for nuclear DNA content by Feulgen microdensitometry according to Greilhuber & Temsch (2001) and Greilhuber (2005). Ten nuclei of mid-prophase cells (4C) were measured per slide and three slides were analysed in total using a Vickers M85a microdensitometer and each nucleus was read three times. Allium cepa L. ‘Ailsa Craig’ (1C = 16.75 pg; Bennett & Smith, 1976) was used as the calibration standard. The 4C-value of each sample was calculated against the 4C-value of the standard in picograms and converted to give the 1C-value.
Results
Alignment of data sets
The ITS data matrix of 56 taxa, three of which were the outgroup, comprised 778 characters, of which 196 were potentially parsimony informative (25.2%). Analysis of ITS sequences yielded 35 equally most-parsimonious trees of 425 steps, consistency index (CI) = 0.82, retention index (RI) = 0.90. One of the most-parsimonious trees was chosen randomly. Tree topology, bootstrap percentages (BP), branches that collapse in the strict consensus tree obtained from maximum parsimony analysis and Bayesian posterior probability values (PP) are indicated in Fig. 1. In the ITS tree, the genus Paphiopedilum is monophyletic, with strong support (100 BP, 1.00 PP). Subgenus Parvisepalum is the first branching clade with 88 BP and 1.00 PP support for monophyly. The support for monophyly of subgenus Brachypetalum was 99 BP and 1.00 PP. Subgenus Paphiopedilum forms a polytomy with subgenus Brachypetalum (60 BP, – PP). Sections Barbata, Pardalopetalum and Cochlopetalum were well supported with 97 BP, 1.00 PP, 100 BP, 1.00 PP and 98 BP, 0.98 PP, respectively. Section Paphiopedilum had moderate bootstrap support (75 BP) but high PP values (0.98). There was no support for section Coryopedilum, and it did not form a clade in the strict consensus tree. In subgenus Paphiopedilum, the relationships within some sections were still not well supported.
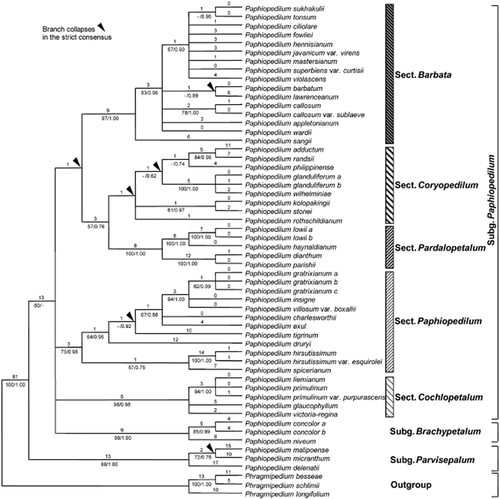
One of 35 most-parsimonious trees from the analysis of the internal transcribed spacer (ITS) region for Paphiopedilum. Tree length = 425, consistency index = 0.82, retention index = 0.90. Numbers above branches are branch lengths and numbers below branches are bootstrap percentages ≥ 50 and posterior probability values ≥ 0.50. Arrows indicate clades that collapse in the strict consensus tree obtained from maximum parsimony analysis. The infrageneric treatment follows Cribb (1998).
The plastid data matrix [partial matK, ycf1, psaA-ycf3ex3 and trnF(GAA)-ndhJ], including 51 taxa (it was not possible to obtain sequences for five taxa that were included in the ITS matrix), three of which were the outgroup, comprised 4353 characters, of which 281 were potentially parsimony informative (6.5%). Analysis of a combined plastid region matrix yielded 20 equally most-parsimonious trees of 520 steps, CI = 0.84, RI = 0.92. One of the most-parsimonious trees was randomly chosen, and the tree topology, bootstrap percentages, branches that collapse in the strict consensus tree obtained from maximum parsimony analysis and Bayesian posterior probability values are indicated in Fig. 2. The tree of the combined plastid regions was more resolved than the ITS tree. The genus Paphiopedilum is monophyletic, with strong support (100 BP, 1.00 PP). The division of the genus into three subgenera is also well supported (100 BP, 1.00 PP for all). Support for the monophyly of Paphiopedilum subgenera Parvisepalum, Brachypetalum and Paphiopedilum is 100 BP, 1.00 PP, 95 BP, 1.00 PP and 100 BP, 1.00 PP, respectively. In subgenus Paphiopedilum, sections Barbata, Paphiopedilum and Pardalopetalum are well supported with 93 BP, 1.00 PP, 98 BP, 1.00 PP and 100 BP, 1.00 PP, respectively. Section Coryopedilum has weak bootstrap support (67 BP) but high PP support (1.00). Section Cochlopetalum forms two clades in a polytomy, with the clade formed by sections Coryopedilum and Pardalopetalum. In subgenus Paphiopedilum, the relationships within some sections are still not well supported.
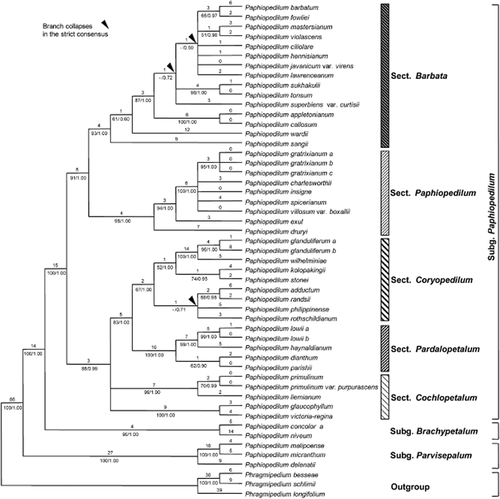
One of 20 most-parsimonious trees from the analysis of plastid (partial matK, ycf1, psaA-ycf3ex3 and trnF(GAA)-ndhJ) regions for Paphiopedilum. Tree length = 520, consistency index = 0.84, retention index = 0.92. Numbers above branches are branch lengths and numbers below branches are bootstrap percentages ≥ 50 and posterior probability values ≥ 0.50. Arrows indicate clades that collapse in the strict consensus tree obtained from maximum parsimony analysis. The infrageneric treatment follows Cribb (1998).
The combined data matrix included 51 taxa (but excluded those for which only ITS data was available), of which three were outgroups, and comprised 4884 characters, of which 463 were potentially parsimony informative (9.5%). Analysis of the combined data matrix yielded 120 equally most-parsimonious trees of 920 steps, CI = 0.83, RI = 0.91. One of the most-parsimonious trees was randomly chosen. Tree topology, bootstrap percentages, branches that collapse in the strict consensus tree obtained from maximum parsimony analysis and Bayesian posterior probability values are indicated in Fig. 3. The genus Paphiopedilum is monophyletic, with strong support (100 BP, 1.00 PP). The division of the genus into three subgenera is well supported (100 BP, 1.00 PP for all). The monophyly of Paphiopedilum subgenera Parvisepalum, Brachypetalum and Paphiopedilum is well supported, with BP 100, 1.00 PP for each node. In subgenus Paphiopedilum, sections Barbata, Paphiopedilum, Pardalopetalum and Cochlopetalum have strong support with 100 BP, 1.00 PP, 99 BP, 1.00 PP, 100 BP, 1.00 PP and 99 BP, 1.00, respectively. Only section Coryopedilum has weak bootstrap support (54 BP) and it collapses to form a polytomy with section Pardalopetalum in the strict consensus; however, it has a high PP value (0.95). In subgenus Paphiopedilum, the relationships within some sections are still not well supported.
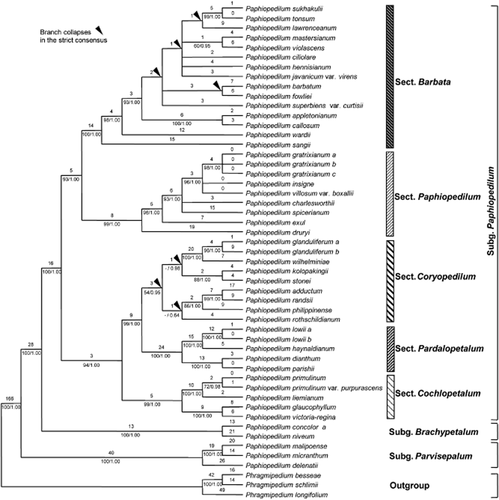
One of 120 most-parsimonious trees from the combined analysis of internal transcribed spacer (ITS) and plastid (partial matK, ycf1, psaA-ycf3ex3 and trnF(GAA)-ndhJ) regions for Paphiopedilum. Tree length = 920, consistency index = 0.83, retention index = 0.91. Numbers above branches are branch lengths and numbers below branches are bootstrap percentages ≥ 50 and posterior probability values ≥ 0.50. Arrows indicate clades that collapse in the strict consensus tree obtained from maximum parsimony analysis. The infrageneric treatment follows Cribb (1998).
Genome size evolution
Genome size data obtained from this study (seven taxa) and from the literature (25 taxa) are listed in Table 3. In Fig. 4, genome size range (1C-value), mean value and chromosome number for each section within the genus are mapped onto the combined tree.
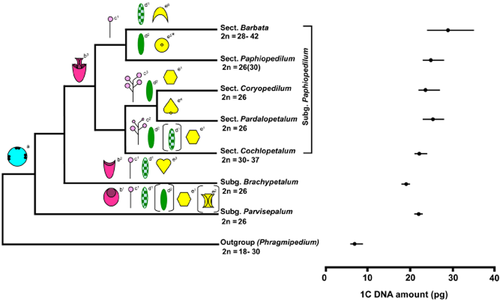
Morphological characters, chromosome numbers and genome size ranges (mean value indicated by a circle) mapped onto a phylogenetic framework from the combined DNA sequence data. a, unilocular ovary with parietal placentation; b1, inflated lip; b2, ovoid shaped lip; b3, lip with only incurved side lobes; c1, (mostly) single-flowered inflorescence; c2, multi-flowered with successive opening; c3, multi-flowered with simultaneous opening; d1, tessellated leaves; d2, plain green leaves; e1, convex staminode; e2 , conduplicate staminode; e3, staminode with uni- or tridentate apex; e4, obcordate staminode with basal protuberance; e5, staminode with an umbo (* indicates more shape variations in the section); e6, (mostly) lunate shape staminode.
| Taxa | Voucher/source | Chromosome number (2n) | 1C-value (pg) |
|---|---|---|---|
| Subgenus Parvisepalum | |||
| Paphiopedilum armeniacum S.C.Chen & F.Y.Liu | Bennett & Leitch, 2010 | 26 | 21.10 |
| Paphiopedilum delenatii Guillaumin | Cox et al., 1998 | 26 | 21.83 |
| Paphiopedilum micranthum Tang & F.T.Wang | Cox et al., 1998 | 26 | 22.75 |
| Subgenus Brachypetalum | |||
| Paphiopedilum concolor (Bateman) Pfitzer | Cox et al., 1998 | 26 | 19.48 |
| Paphiopedilum godefroyae (God.-Leb.) Stein | Cox et al., 1998 | 26 | 17.80 |
| Subgenus Paphiopedilum | |||
| Section Paphiopedilum | |||
| Paphiopedilum insigne (Wall. ex Lindl.) Pfitzer | Kew 2001–2843 | 26 | 27.52 (0.59)a |
| Paphiopedilum gratrixianum (Mast.) Rolfe | Kew 1979–975 | 26 | 25.16 (0.46)a |
| Paphiopedilum druryi (Bedd.) Stein | Kew 1982–1398 | 30 | 26.50 (0.47)a |
| Paphiopedilum villosum (Lindl.) Stein | Narayan et al., 1989 | 26 | 22.48 |
| Section Barbata | |||
| Paphiopedilum appletonianum (Gower) Rolfe | Cox et al., 1998 | 38 | 32.43 |
| Paphiopedilum mastersianum (Rchb.f.) Stein | Cox et al., 1998 | 36 | 29.73 |
| Paphiopedilum tonsum (Rchb.f.) Stein | Cox et al., 1998 | 32 | 28.15 |
| Paphiopedilum barbatum (Lindl.) Pfitzer | Cox et al., 1998 | 38 | 33.75 |
| Paphiopedilum bullenianum (Rchb.f.) Pfitzer var. celebesense (Fowlie & Birk) P.J.Cribb | Bennett & Leitch, 2010 | 40 | 25.85 |
| Paphiopedilum callosum (Rchb.f.) Stein | Cox et al., 1998 | 32 | 24.05 |
| Paphiopedilum lawrenceanum (Rchb.f.) Pfitzer | Bennett & Leitch, 2010 | 40 | 26.13 |
| Paphiopedilum ciliolare (Rchb.f.) Stein | Bennett & Leitch, 2010 | 32 | 30.50 |
| Paphiopedilum purpuratum (Lindl.) Stein | Bennett & Leitch, 2010 | 40 | 27.13 |
| Paphiopedilum sukhakulii Schoser & Senghas | Cox et al., 1998 | 40 | 29.73 |
| Paphiopedilum wardii Summerh. | Cox et al., 1998 | 41 | 34.53 |
| Section Pardalopetalum | |||
| Paphiopedilum parishii (Rchb.f.) Stein | Kew 1986–1038 | 26 | 27.20 (0.68)a |
| Paphiopedilum lowii (Lindl.) Stein | Bennett & Leitch, 2010 | 26 | 24.53 |
| Paphiopedilum haynaldianum (Rchb.f.) Stein | Bennett & Leitch, 2010 | 26 | 22.85 |
| Section Cochlopetalum | |||
| Paphiopedilum liemianum (Fowlie) K.Karas. & K.Saito | Kew 1990–8000 | 32 | 23.72 (0.48)a |
| Paphiopedilum primulinum M.W.Wood & P.Taylor | Cox et al., 1998 | 32 | 20.90 |
| Paphiopedilum victoria-mariae (Sander ex Mast.) Rolfe | Cox et al., 1998 | 36 | 21.40 |
| Section Coryopedilum | |||
| Paphiopedilum philippinense (Rchb.f.) Stein | Cox et al., 1998 | 26 | 23.25 |
| Paphiopedilum kolopakingii Fowlie | Kew 1983–5478 | 26 | 21.93 (0.86)a |
| Paphiopedilum stonei (Hook.) Stein | Kew 1998–2185 | 26 | 23.28 (0.46)a |
| Paphiopedilum adductum Asher | Bennett & Leitch, 2010 | 26 | 27.03 |
| Paphiopedilum glanduliferum (Blume) Stein | Cox et al., 1998 | 26 | 23.73 |
| Paphiopedilum rothschildianum (Rchb.f.) Stein | Cox et al., 1998 | 26 | 22.58 |
| Outgroup | |||
| Phragmipedium besseae Dodson & J.Kuhn | Cox et al., 1998 | 24 | 7.08 |
| Phragmipedium longifolium (Warsz. & Rchb.f.) Rolfe | Cox et al., 1998 | 20, 21, 22, 23 | 6.10 |
| Phragmipedium caudatum (Lindl.) Rolfe | Cox et al., 1998 | 28 | 9.18 |
| Phragmipedium lindleyanum (R.H.Schomb. ex Lindl.) Rolfe | Cox et al., 1998 | 22 | 8.03 |
| Phragmipedium pearcei (Rchb.f.) Rauh & Senghas | Cox et al., 1998 | 20, 21, 22 | 6.33 |
- a Standard deviations of 1C-values measured in this study are shown in parentheses (pg).
Discussion
Congruence of ITS and plastid data
The results from two separate matrices of ITS and plastid data showed no conflict between strongly supported branches (> 75 BP, > 0.90 PP) when compared node by node. Groupings in the genus in both ITS and plastid trees are generally as described in the treatment of Cribb (1998), but the relationships along the backbone are less resolved in the ITS tree. The results in the plastid trees had better bootstrap support, but the resulting trees from separate analyses of each individual plastid region [partial matK, ycf1, psaA-ycf3ex3 and trnF(GAA)-ndhJ] lacked resolution because of low levels of divergence (data not shown). The combined data set produced more resolved trees, mostly with strong bootstrap support. In general the increase in clade support in the combined tree (Fig. 3) indicates congruence between the ITS and plastid data. The only place where there was lower clade support when the plastid and nuclear data sets were combined was in section Coryopedilum, suggesting some possible conflict between data sets in this part of the phylogenetic tree. However, the branches concerned receive only low bootstrap support.
Phylogenetic relationships in the genus Paphiopedilum
Overall, the results from all analyses showed general congruence with the previous infrageneric treatment of Cribb (1998), and confirm that the genus Paphiopedilum is monophyletic, which is congruent with the results of previous studies (Albert, 1994; Cox et al., 1997).
Subgenus Parvisepalum
Subgenus Parvisepalum, characterized by tessellated leaves (except two species, P. hangianum and P. emersonii, which have plain green leaves; Averyanov et al., 2003), a single-flowered inflorescence, a flower with an inflated lip and a convex (mostly) or conduplicate staminode (Cribb, 1998) (Fig. 4), was found to be the first branching clade with strong support in this study (Figs 2, 3). This confirms the results of Cox et al. (1997) and the suggestion of Chen & Tsi (1984) that P. malipoense S.C.Chen & Z.H.Tsi and its closely related species are the ‘basal group’ (i.e. early diverging) of the genus. Chen & Tsi (1984) suggested that Paphiopedilum and Cypripedium were related via this species (subgenus Parvisepalum) by considering the similarity of the flower characters. However, Cribb (1987) stated that the similarities between the flowers of Paphiopedilum and the other genera, for example P. armeniacum S.C.Chen & F.Y.Liu and C. irapeanum La Llave & Lex. or P. delenatii Guillaumin and Phragmipedium schlimii (Linden ex Rchb.f.) Rolfe, are the result of similar pollination syndromes with bees as pollinators. Research into seven species in subgenus Paphiopedilum and one species in subgenus Brachypetalum showed that all of them are pollinated by hoverflies (Atwood, 1985; Bänziger, 1994, 1996, 2002; Shi et al., 2007, 2009), but there is no such research for species in subgenus Parvisepalum. The results from the studies of Albert (1994) and Cox et al. (1997) pointed to Paphiopedilum differing extensively from both Cypripedium and Phragmipedium, not only in morphological characters but also in molecular characters. In this study, the results from the combined data of five DNA regions also showed that there are high levels of molecular divergence between Paphiopedilum and Phragmipedium.
Subgenus Brachypetalum
Subgenus Brachypetalum, characterized by tessellated leaves, one- or two- (rarely three-) flowered inflorescences, flowers white or yellow in colour, an involute margined ovoid shaped lip and a staminode that is uni- or tridentate at its apex (Cribb, 1998) (Fig. 4), is a monophyletic group, with high support values from both BP and PP in all analyses. From plastid and combined data (Figs 2, 3), subgenus Brachypetalum is strongly supported as sister to subgenus Paphiopedilum. This result supports the recognition of subgenus Parvisepalum by Karasawa & Saito (1982), which was found to differ morphologically from the remaining species in subgenus Brachypetalum, and the elevation of section Parvisepalum sensu Cribb (1987) to subgeneric level in the second edition of his monograph (Cribb, 1998), a change suggested by the ITS result of Cox et al. (1997). Although both Parvisepalum (most species) and Brachypetalum have tessellated leaves and a sporophytic chromosome number of 26, their flowers are clearly different (Fig. 4). Approximately seven species of subgenus Parvisepalum are distributed mostly in southern China and Vietnam, whereas the four species of Brachypetalum have a wider distribution in mainland south-east Asia (Cribb, 1998).
Subgenus Paphiopedilum
There is conflict between the classical infrageneric classifications concerning the division of subgenus Paphiopedilum into several sections or several subgenera in the most recent monographs of the genus. In the monographs of Braem (Braem, 1988; Braem et al., 1998; Braem & Chiron, 2003), following the work of Karasawa & Saito (1982), subgenus Paphiopedilum sensu Cribb is divided into four subgenera (Paphiopedilum, Sigmatopetalum Hallier f. ex K.Karas. & K.Saito, Polyantha (Pfitzer) Brieger and Cochlopetalum (Hallier f. ex Pfitzer) K.Karas. & K.Saito). This disagrees with the treatment of Cribb in his monographs (Cribb, 1987, 1998), in which he placed plants with different leaf colour (plain green vs. tessellated), number of flowers in the inflorescence [one or rarely two (three) flowers vs. multiple flowers], number of chromosomes (constant 2n = 26 vs. variable) and pattern of blooming (simultaneous vs. successive), in one subgenus (Braem & Chiron, 2003). However, Cribb considered subgenus Paphiopedilum to be monophyletic based on the cladistic study of Atwood (1984) and he treated other groups at sectional levels in this subgenus. Braem (in Braem & Chiron, 2003) also argued that the ITS tree from Cox et al. (1997) did not disagree with his subgeneric treatment. That is because there is no support for the robustness of the clade of subgenus Paphiopedilum sensu Cribb, as mentioned previously.
The results from this study show that subgenus Paphiopedilum sensu Cribb which consists of species in which only the side lobes of the lip are incurved (Cribb, 1998) (Fig. 4), is clearly monophyletic, with strong support from the plastid and combined data analyses (Figs 2, 3), and the subgenus is split into two main lineages. The first lineage includes three sections of multi-flowered species (Coryopedilum, Pardalopetalum and Cochlopetalum) and the second lineage includes two sections of mostly single-flowered species (Paphiopedilum and Barbata) (Figs 2-4). These are all sections as defined in the treatment of Cribb (1998). These lineages are different from the results of Cox et al. (1997), in which multi-flowered and (mostly) single-flowered sections are placed in the same clades. In the current study, multi-flowered inflorescences occur only in sections Coryopedilum, Pardalopetalum and Cochlopetalum, and thus this character appears to be a synapomorphy for this clade.
The tessellated leaf character found in the early diverging subgenera Parvisepalum (except two species) and Brachypetalum, is absent in most clades of subgenus Paphiopedilum (Fig. 4). Reversions of this character are found in all species of section Barbata and in two species of section Cochlopetalum, and it appears to occur independently. Tessellated leaves are thought to play a role as camouflage for anti-herbivore defence in understorey herbaceous plants growing in sun-flecked light conditions (Givnish, 1990), but there is no obvious evidence for the value of this adaptation in Paphiopedilum. Most species, including those with plain green and tessellated leaves, grow in similar shady forest-floor habitats, although a few plain green leaved species have been found in open sunny situations and some tessellated leaved species are found in deep shade (Cribb, 1998).
All sections in subgenus Paphiopedilum are strongly supported (both BP and PP) in the analyses of combined data, except section Coryopedilum, which has weak BP support (54 BP) for monophyly, collapsing in the strict consensus tree of parsimony analysis to form a polytomy with section Pardalopetalum. However, in the tree obtained from Bayesian analysis, Coryopedilum has 0.95 PP clade support (Fig. 3). Previously, the results from ITS data of Cox et al. (1997) showed section Coryopedilum (no BP support, jackknife > 0.63 at some nodes) to be paraphyletic to a monophyletic section Pardalopetalum sensu Cribb (1987), and they tentatively proposed a combination of these sections. However, Cribb (1998), in the second edition of his monograph, did not accept these molecular results, because he noted that these sections are probably sister groups based on morphological characters. The sections share plain green leaves, multi-flowered inflorescences that open simultaneously and a chromosome number of 2n = 26 (Fig. 4). Considering floral morphology, they can be clearly distinguished, with Coryopedilum having long tapering petals, a porrect lip and a convex staminode, whereas Pardalopetalum has distinctive dorsal petals that are reflexed at the base and an obcordate staminode with a basal protuberance and tridentate apex (Cribb, 1998). The c. 11 species of section Coryopedilum are found in the Malesian islands, and most are endemic to single islands. In contrast, section Pardalopetalum is more widespread, the four species being distributed through mainland south-east Asia, and the Malay Archipelago to Sulawesi and the Philippines (Cribb, 1998). In this study (Figs 1-3), these sections are sister groups, with 57 BP and 0.76 PP from ITS data, 83 BP and 1.00 PP from the plastid data and 99 BP and 1.00 PP from the combined data. There is no support for monophyly from the ITS data for Coryopedilum. Although, bootstrap support from plastid data and combined data is low (67 BP and 54 BP, respectively), support from Bayesian analysis is high, with 1.00 PP from plastid data and 0.95 PP from the combined data. However, Coryopedilum collapsed in the strict consensus trees of parsimony analyses of ITS data and combined data. In contrast, section Pardalopetalum has strong support, with 100 BP and 1.00 PP in all analyses. Results from this study therefore suggest that section Coryopedilum, although clearly differing from section Pardalopetalum morphologically, shows insufficient levels of molecular divergence to support monophyly of this section. Including more variable regions such as low-copy nuclear regions would possibly help in obtaining a clearer pattern. The low level of molecular divergence in Coryopedilum could possibly be explained by its selfing mode of reproduction, resulting from geitonogamy, and an absence of centric fission events (see below). Species with multi-flowered inflorescences that open simultaneously, as found in sections Coryopedilum and Pardalopetalum, are more susceptible to geitonogamy or pollination among flowers on the same individual plant (Kliber & Eckert, 2004). This self-pollination by geitonogamy is thought to be disadvantageous, because it produces inbred offspring and requires pollinators to visit, as in outcrossing pollination (Eckert, 2000). Although the floral features of orchids favour outcrossing, most orchids are self-compatible, which could facilitate reproduction in widely separated plants where outcrossing is not possible (Dressler, 1981). Because most species in section Coryopedilum are endemic to single Malesian islands (Cribb, 1998), they occur in small populations that are more likely to be geitonogamous than those of species in section Pardalopetalum, which are distributed more widely.
The Cochlopetalum clade is recovered in trees from ITS data (98 BP and 0.98 PP) and combined data (99 PP and 1.00 PP), but not in the plastid tree. In the combined tree, section Cochlopetalum is sister to a clade formed by sections Coryopedilum and Pardalopetalum (94 BP and 1.00 PP). Section Cochlopetalum is similar to its sister group in having multi-flowered inflorescences, but it differs in its flowers, which open successively, and in the variation in chromosome numbers (2n = 30–37) (Fig. 4). In addition, linear, spirally twisted petals are a distinctive character for the section, including approximately five species that are endemic to Java and Sumatra (Cribb, 1998). These three sections, which share plain green leaves [except P. victoria-regina (Sander) M.W.Wood and P. victoria-mariae (Sander ex Mast.) Rolfe of section Cochlopetalum, which have faintly tessellated leaves; Cribb, 1998] and multi-flowered inflorescences, are together sister to a clade consisting of sections Paphiopedilum plus Barbata with strong support (100 BP and 1.00 PP from both plastid and combined data). The clade of sections Paphiopedilum and Barbata is characterized by single-flowered (rarely two-flowered) inflorescences (Fig. 4). Both sections are monophyletic with strong support: 98 BP and 1.00 PP from plastid data, and 99 BP and 1.00 PP from combined data for Paphiopedilum; 93 BP and 1.00 PP from plastid data; and 100 BP and 1.00 PP from a combined data for Barbata (Figs 2, 3). Section Paphiopedilum differs from section Barbata in having green leaves and chromosome numbers in most species of 2n = 26 except in P. druryi (Bedd.) Stein and P. spicerianum (Rchb.f.) Pfitzer (2n = 30), whereas the tessellated-leaved section, Barbata shows considerable variation in chromosome number (2n = 28–42). Many species in section Paphiopedilum are characterized by a staminode with an umbo in the middle, whereas most species in section Barbata have a lunate staminode (Cribb, 1998) (Fig. 4).
Phylogenetic relationships in section Barbata are unresolved, with many internal branches collapsing to a polytomy in the strict consensus tree for the parsimony analysis and 50% majority tree from Bayesian analyses (Figs 1-3). Atwood (1984) suggested that section Barbata was the most derived group, and this section was derived from section Paphiopedilum based on his Wagner groundplan-divergence cladogram. However, that suggestion cannot be inferred from this current phylogenetic study, because it can only be inferred that both sections share a most recent common ancestor. The short branch lengths in section Barbata shown on the combined tree in this study and the narrow geographical distribution on Malesian islands of most species in this section might suggest a recent rapid radiation in the section (Cox et al., 1997). Although we included numerous molecular characters from five DNA regions both from nuclear and plastid loci in this study, the relationships in this section remain unresolved. To obtain better resolution in this section, the use of more variable regions such as low-copy nuclear sequences could be helpful.
Genome size and chromosome evolution in the genus Paphiopedilum
Mapping chromosome number data onto the phylogenetic framework from the combined data does not show clearly if there is a trend towards an increase in chromosome number, as proposed by Cox et al. (1997, 1998) (Fig. 4). There are two major lineages in subgenus Paphiopedilum, the first lineage composed of three sections (Coryopedilum, Pardalopetalum and Cochlopetalum). All species in the first two sections of this clade have a chromosome number of 2n = 26, whereas species of section Cochlopetalum have chromosome numbers that vary from 2n = 30 to 2n = 37. Similarly, in the second lineage, species of section Paphiopedilum have a chromosome number of 26 (except two species, P. druryi and P. spicerianum, with 2n = 30), whereas variable chromosome numbers, between 2n = 28 and 42, are found in the sister section Barbata. Although the topology of sections in subgenus Paphiopedilum in this phylogenetic framework is different from the study of Cox et al. (1997, 1998), the patterns are similar, in that sections with variable chromosome numbers are paired with sections with a constant chromosome number. However, it has been shown from both phylogenetic frameworks that the first branching subgenus, Parvisepalum, and subgenus Brachypetalum, which is sister to subgenus Paphiopedilum, have a chromosome number of 2n = 26, with all metacentric chromosomes, and this could indicate that 2n = 26 is the ancestral condition for the genus, as suggested previously, because this number is found in most species of the genus (e.g. Karasawa, 1979). Also, the higher chromosome number and the presence of telocentric chromosomes could indicate a more derived condition given the phylogenetic position of species with higher chromosome numbers. These results suggest that centric fission has contributed to the karyotype changes observed in the genus and, superimposing the data onto the phylogenetic tree, indicate that centric fission has occurred independently in sections Barbata and Cochlopetalum (Fig. 4).
There have been other studies that support a hypothesis of centric fission, for example Karasawa & Tanaka (1980), who studied C-banding patterns of P. callosum (Rchb.f.) Stein (2n = 32) and found them to be similar to P. insigne (Wall. ex Lindl.) Pfitzer [=P. insigne (Wall. ex Lindl.) Pfitzer var. sanderae (Rchb.f.) Pfitzer, 2n = 26]. They postulated centric fission as a cause of karyotype changes.
Jones (1998), in a review of Robertsonian change in karyotype evolution, supported the hypothesis of centric fission in Paphiopedilum. He suggested that the small population sizes and inbreeding in Paphiopedilum could contribute to explaining the karyotype variation observed. Indeed, all species of section Cochlopetalum and most species of section Barbata that have a high chromosome number are endemic to the Malesian islands, and it has been suggested that centric fission may be under selection as it has the potential to increase genetic recombination, enabling adaptation to the environments on islands (Cox et al., 1998; Leitch et al., 2009). However, this is clearly not always the case, as species of section Coryopedilum, most of which are also restricted to individual Malesian islands (Cribb, 1998), all have a chromosome number of 2n = 26. Although Cox (in Pridgeon et al., 1999) suggested that the higher chromosome number of 2n = 30 in P. druryi (section Paphiopedilum) might be correlated with its narrow endemicity (in southern India), clearly, other factors are involved in driving centric fission. This is because the only other species in section Paphiopedilum with 2n = 30 is P. spicerianum, which has a wider distribution. It is found in north-east India, north-west Burma and south-west China (Cribb, 1998).
The range in genome size, as represented by 32 species (44% of the genus), is from 1C = 17.80 pg in P. godefroyae to 1C = 34.53 pg in P. wardii (1.9-fold range; see Tables 3, 4 and Fig. 4). The lowest genome sizes are found in species belonging to section Brachypetalum (mean 1C = 18.64 pg) and the highest genome sizes are found in section Barbata (mean 1C = 29.27 pg). Mapping the genome size range of Paphiopedilum spp. onto the phylogenetic framework obtained in this study shows that there is no clear trend of genome size increase in the genus (Fig. 4). The greatest range and largest genomes were found in section Barbata, which is also characterized by being the most variable in terms of chromosome number (2n = 28–42). However, section Cochlopetalum, which also is variable in chromosome number (2n = 30–37), has a similar range of genome size to other sections and subgenera characterized by 2n = 26 (Table 4).
| Taxa | Chromosome number (2n) | n.f. | Min. 1C-value (pg) | Max. 1C-value (pg) | Mean 1C-value (pg) | No. species with 1C-value | Representation (%) |
|---|---|---|---|---|---|---|---|
| Subgenus Parvisepalum | 26 | 52 | 21.10 | 22.75 | 21.89 | 3 | 43 |
| Subgenus Brachypetalum | 26 | 52 | 17.80 | 19.48 | 18.64 | 2 | 50 |
| Subgenus Paphiopedilum | |||||||
| Section Cochlopetalum | 30-37 | 48–50 | 20.90 | 23.72 | 22.01 | 3 | 60 |
| Section Pardalopetalum | 26 | 52 | 22.85 | 27.20 | 24.86 | 3 | 75 |
| Section Coryopedilum | 26 | 52 | 21.93 | 27.03 | 23.63 | 6 | 55 |
| Section Paphiopedilum | 26 (30)a | 52 | 22.48 | 27.52 | 25.42 | 4 | 29 |
| Section Barbata | 28–42 | 52–56 | 24.05 | 34.53 | 29.27 | 11 | 41 |
| Phragmipedium (outgroup) | 18–30 | 34–39 | 6.10 | 9.18 | 7.34 | 5 | 33 |
- a P. druryi and P. spicerianum 2n = 30.
When plotting chromosome number against genome size data (Fig. 5), a weak but significant relationship was found (Pearson's correlation coefficient r = 0.632, P < 0.001), suggesting that, as chromosomes undergo fission, this is often accompanied by an increase in genome size. The source of additional DNA in the genome is unclear, but is likely to comprise a diverse array of different types of repetitive DNA, including retrotransposons (Bennetzen, 2005).
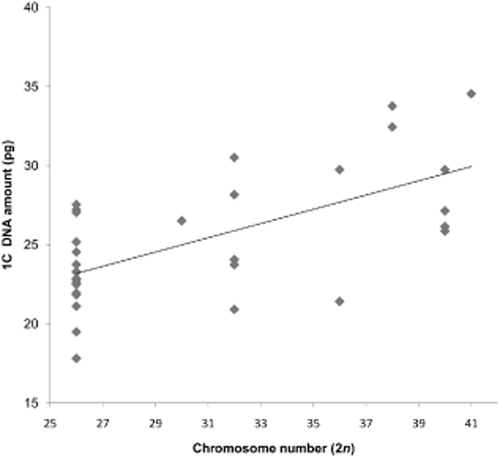
The relationship between genome size and chromosome number for 32 Paphiopedilum spp. Pearson's correlation coefficient r = 0.632, P < 0.001.
The relationship between chromosome number and genome size in Paphiopedilum differs from that of closely related genera. Phragmipedium has a variable chromosome number (2n = 18–30), but a smaller mean genome size and a narrower range (1.5-fold, 1C = 6.10 to 9.18 pg) (Cox et al., 1998). Cypripedium is the most variable genus in subfamily Cypripedioideae in terms of genome size, with values ranging 10.5-fold (1C = 4.1 to 43.1 pg), but the chromosome number in most species is constant (2n = 20) (Leitch et al., 2009).
Lan & Albert (2011) studied the evolution of ribosomal DNA in Paphiopedilum using fluorescence in situ hybridization and assessed the data according to the phylogenetic framework of Cox et al. (1997). Although the results show variation of rDNA multiplication in Paphiopedilum, they found no evidence for a clear relationship between the increase in number of chromosomal locations of rDNA and the increase in chromosome number and genome size. Using the more robust phylogenetic framework from the current study, the multiplication of 25S rDNA loci observed by Lan & Albert occurred twice independently in Paphiopedilum, once in subgenus Parvisepalum and once in the clade formed by sections Coryopedilum and Pardalopetalum of subgenus Paphiopedilum. The multiplication event of 5S rDNA loci happened only in subgenus Paphiopedilum, whereas the early diverging subgenera Parvisepalum and Brachypetalum retained the ancestral number of two major sites, as also found in the outgroups Phragmipedium and Mexipedium.
Genome size is thought to have an influence on life form, habit and ecology. Annual plants are characterized by small genomes, whereas perennials have a larger range of genome sizes, and species with large genomes are all obligate perennials (Bennett, 1972). Leitch et al. (2009) found that epiphytic orchids have small genomes (mean 1C = 3.0 pg, range 0.33–8.5 pg), whereas terrestrial species have a much wider range (mean 1C = 18.3 pg, range 2.9–55.4 pg). This might be caused by selection for small guard cell sizes, because species with small guard cells are shown to respond more rapidly to water stress than those with larger cells (Aasamaa, Sober & Rahi, 2001; Hetherington & Woodward, 2003). As guard cell size has been shown to be correlated with genome size, then selection for small guard cells would result in selection for a small genome (Beaulieu et al., 2008). Most Paphiopedilum spp. are terrestrials, with only five being epiphytic; P. parishii (Rchb.f.) Stein, P. lowii (Lindl.) Stein, P. villosum (Lindl.) Stein, P. hirsutissimum (Lindl. ex Hook.) Stein and P. glanduliferum (Blume) Stein, the last two species being facultative epiphytes (Cribb, 1998). Nevertheless, in contrast to the observations of Leitch et al. (2009), the genome size of these epiphytic species is large (mean 1C = 24.49 pg, range 22.48–27.20 pg), similar to those found in the terrestrial species (mean 1C = 25.40 pg, range 17.80–34.53 pg). These observations suggest that water stress is unlikely to be a strong selective pressure on cell size in this case, perhaps because the high rainfall in habitats where Paphiopedilum spp. are found is seasonal. In addition, other features, such as thick leathery leaves, could also be strategies that enable their survival in the dry season (Cribb, 1998).
Acknowledgements
We thank Dr J. Clarkson, Dr D. Devey, Dr L. Lledó, R. Cowan, L. Csiba and E. Kapinos (Jodrell Laboratory, RBG, Kew) for their technical support and advice, and thanks especially to J. Joseph, M. Sanchez, Dr H. Saslis-Lagudakis, Dr J. Pellicer, Dr L. Kelly, Dr R. Kynast, Dr I. Kahandawala and Dr M. Zarrei for their advice, support and valuable help. Thanks also to C. Ryan and B. Kompalli (The Tropical Nursery RBG, Kew) for giving access to living plant collections and to anonymous reviewers for their valuable comments on this manuscript.




