Urinary biomarkers of prostate cancer
Abstract
The development of more specific biomarkers for prostate cancer and/or high-risk prostate cancer is necessary, because the prostate-specific antigen test lacks specificity for the detection of prostate cancer and can lead to unnecessary prostate biopsies. Urine is a promising source for the development of new biomarkers of prostate cancer. Biomarkers derived from prostate cancer cells are released into prostatic fluids and then into urine. Urine after manipulation of the prostate is enriched with prostate cancer biomarkers, which include prostate cancer cells, DNAs, RNAs, proteins and other small molecules. The urinary prostate cancer antigen 3 test is the first Food and Drug Administration-approved RNA-based urinary marker, and it helps in the detection of prostate cancer on repeat biopsy. The SelectMDx test is based on messenger RNA detection of DLX1 and HOXC6 in urine after prostate massage, and helps in the detection of high-risk prostate cancer on prostate biopsy. Exosomes are extracellular vesicles with a diameter of 30–200 nm that are secreted from various types of cells. Urinary prostate cancer-derived exosomes also contain RNAs and proteins specific for prostate cancer (e.g. PCA3 and TMPRSS2-ERG), and could be promising sources of novel biomarker discovery. The ExoDx Prostate test is a commercially available test based on the detection of three genes (PCA3, ERG and SPDEF) in urinary exosomes. Advancement of comprehensive analysis (microarray, mass spectrometry and next-generation sequencing) has resulted in the discovery of several urinary biomarkers. Non-invasive urinary markers can help in the decision to carry out prostate biopsy or in the design of a therapeutic strategy.
Abbreviations & Acronyms
-
- 5-ALA
-
- 5-aminolevulinic acid
-
- ADIRF
-
- adipogenesis regulatory factor
-
- AMACR
-
- α-methylacyl-CoA racemase
-
- AR
-
- androgen receptor
-
- AUC
-
- area under the curve
-
- BPH
-
- benign prostatic hyperplasia
-
- CRPC
-
- castration-resistant prostate cancer
-
- DRE
-
- digital rectal examination
-
- EMT
-
- epithelial-mesenchymal transition
-
- ETS
-
- E-twenty six
-
- EV
-
- extracellular vesicle
-
- GCNT1
-
- core 2 β-1,6-N-acetylglucosaminyltransferase-1
-
- GSTP1
-
- S-transferase gene
-
- HOXC6
-
- homeobox C6
-
- iTRAQ
-
- isobaric tags for relative and absolute quantification
-
- LC-MS/MS
-
- liquid chromatography tandem-mass spectrometry
-
- mRNA
-
- messenger RNA
-
- miR
-
- microRNA
-
- NA
-
- not available
-
- PCA3
-
- prostate cancer antigen 3
-
- PGA3
-
- pepsinogen 3, group 1
-
- PPIX
-
- protoporphyrin IX
-
- PSA
-
- prostate-specific antigen
-
- THP
-
- Tamm Horsfall protein
-
- TMPRSS2
-
- transmembrane protease serine 2 gene
-
- ZAG
-
- zinc-alpha2-glycoprotein
-
- β2M
-
- β-2-microglobulin
Introduction
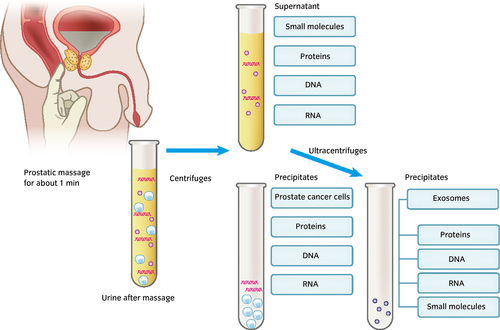
The incidence rate of prostate cancer is increasing, and is highest in Japan and the USA. PSA is the gold standard test for the screening and diagnosis of prostate cancer. However, PSA levels are also elevated in men with BPH or prostatic inflammation. The specificity of the PSA test in men with a PSA level of 4.0–10.0 ng/mL is approximately 20–45%, and a number of men with elevated PSA levels have experienced an unnecessary prostate biopsy, which carries significant morbidity.1 The PSA test cannot differentiate high-risk prostate cancer, resulting in a recommendation against the PSA screening test by the US Preventive Services Task Force. Therefore, the development of more specific biomarkers for prostate cancer and/or high-risk prostate cancer is necessary. Urine is a promising source for the development of new biomarkers of prostate cancer. Prostate cancer cells or prostatic intraepithelial neoplasia arising from the prostate epithelium can secrete substances or release their cellular contents into the prostatic glands, and prostate cancer cells themselves can be shed into the prostatic glands. The prostatic fluids containing substances derived from prostate cancer cells are exuded into the urine after manipulation of the prostate by DRE (Fig. 1). Some of these substances can be detected in urine even without prostate manipulation by a sensitive assay or if they exist in urine in a large amount. For example, PSAs existing in the prostatic fluids exudate in urine and can be detected in urine without prostate manipulation. Prostatic manipulation might annoy patients, but could increase the level of biomarkers in the urine. Biomarker candidates, such as prostate cancer cells, DNA, RNA, proteins, exosomes and other small molecules, exist in urine.2 Centrifugation of the first 10–50 mL of urine voided after prostate massage at 1000–2000 g results in precipitates of urine that can contain prostate cancer cells and fragments of prostate cancer cells, which also contain DNA, RNA and proteins derived from these cells. Supernatants contain free proteins, DNAs, RNAs, other small molecules and exosomes. Further ultracentrifugation of the supernatant results in the precipitation of exosomes (Fig. 1). Recent progress in new techniques is enabling us to develop various new urinary markers of prostate cancer (Table 1).
| Urine | Sensitivity | Specificity | AUC | Reference | ||
|---|---|---|---|---|---|---|
| Prostate cancer cells | ||||||
| Multiple immunostaining against AMACR, Nkx3.1 and nucleolin | After massage | Precipitate | 36% | 100% | 5 | |
| 5-ALA | After massage | Precipitate | 74% | 70% | 6 | |
| Proteins | ||||||
|
β-2-microglobulin Pepsinogen 3, group 1 Intestinal mucin |
No manipulation | Whole urine |
0.66 0.62 0.61 |
9 | ||
| Endoglin | After massage | Supernatant | 73% | 62% | 0.72 | 10 |
| AMCR | After massage | Precipitate | 100% | 58% | 12 | |
| IL18-Bpa | After massage | Supernatant | 69% | 56% | 0.65 | 13 |
| Annexin A3 | After massage | NA | 0.82 | 14 | ||
| Engrailed-2 | No manipulation | Supernatant | 66% | 88% | 15 | |
| ZAG | After massage | Supernatant | 0.75 | 16 | ||
| Core2 β-1,6-N-acetylglucosaminyltransferase | After massage | Supernatant | 0.76 for the prediction of extracapsular extension | 17 | ||
| Urinary glycosylation profile | After massage | Supernatant | 0.84 | 18 | ||
| Fucosylated PSA | After massage | Supernatant | 77% | 80% | 20 | |
| DNA methylation | ||||||
| GSTP1 promoter methylation | After massage | Precipitate | 73% | 98% | 22 | |
| RASF1 promoter methylation | After massage | Precipitate | 45% | 84% | 26 | |
|
APC methylation RAR beta2 methylation GSTP1 methylation |
After massage | Precipitate |
0.58 0.70 0.66 |
27 | ||
|
HIST1H4K methylation GSTP1 methylation |
After massage | Precipitate |
43 45 |
80 80 |
0.64 0.69 |
29 |
| 4-gene methylation classifier panel (APC, CRIP3, GSTP1 and HOXD8) | After massage | Precipitate | Prediction of the patients’ reclassification on the active surveillance | 31 | ||
| Cell-free DNA | ||||||
|
AR amplification TMPRSS2-ERG fusion PTEN deletion NOTCH1 locus amplification MYCL amplification |
No manipulation | Supernatant | Detectable in 5 of 10 CRPC patients | 32 | ||
| c-Myc, HER2 and AR | No manipulation | Supernatant | 0.50 | 33 | ||
| RNA | ||||||
| PCA3 | After massage |
Precipitate Whole urine Whole urine |
67 65 |
73 |
0.75 | |
| TMPRSS2-ERG, PCA3 | After massage | Whole urine | 93% for aggressive PCa | 33% for aggressive PCa | 46 | |
| HOXC6, TDRD1 and DLX16 | After massage | Precipitate | 0.77 for aggressive PCa | 48 | ||
| HOXC6, DLX1 and KLK3 | After massage | 0.76 for aggressive PCa | 49 | |||
| MicroRNA | ||||||
|
miR-107 miR-574-3p |
NA | Precipitate |
0.74 0.66 |
52 | ||
| miR-205 and miR-214 | NA | NA | 89% | 80% | 53 | |
|
miR-1825 miR-484 |
NA | NA |
60% 80% |
69% 19% |
54 | |
| miR-183 and miR-205 | After massage | Precipitate | 90% | 3% | 55 | |
| Volatile small molecule | ||||||
| By olfactory systems of a well-trained canine | No manipulation | Whole urine | 100% | 98% | 71 | |
| 2,6-dimethyl-7-octen-2-ol, pentanal, 3-octa- none and 2-octanone, | No manipulation | Whole urine | 80% | 57% | 0.76 | 72 |
Prostate cancer cells in urine
It has been known for almost 70 years that prostate cancer cells are shed into prostatic fluids by manipulation of the prostate.3 Cytological examination is a reliable test for the detection of urothelial carcinoma with high specificity. In a similar manner, cytological examination was also assessed for its ability to detect prostate cancer, but it failed due to its low sensitivity.4 Urine cytological examination can diagnose the cells as malignant, but it is difficult to determine their origin; that is, whether the malignant cells originate from urothelial carcinoma or prostate cancer. Multiple staining of the precipitates of urine after prostate massage for AMACR, Nkx3.1, nucleolin and 4′-6-diamidino-2-phenylindole to detect prostate cancer cells in urine was also carried out.5 Upregulation of AMACR and downregulation of Nkx3.1 were specific for prostate cancer, and the upregulation of nucleolin was a characteristic of the malignant cells. Multiplex immunofluorescence cytology was compared with the conventional cytological examination for prostate cancer detection. Among 50 men who underwent prostate biopsy, the precipitates of urine after prostate massage were stained, and the sensitivity for prostate cancer detection was 36% (9/25) and the positive predictive value was 100% (8/8). This multiplex immunofluorescence study was the first report to prove that malignant cells in urine after massage originated from prostate cancer. Typically, small clumps of prostate cancer cells were shed into the prostatic fluid. The new technique of photodynamic diagnosis was also applied for the detection of prostate cancer cells in urine after massage.6 Administration of the photodynamic agent 5-ALA induces PPIX accumulation in malignant tissue, which then enables its differentiation from benign tissue.7 Urine after massage was obtained from 138 men with elevated PSA or abnormal DRE findings before biopsy. Urinary precipitates after centrifugation were treated with 5-ALA and imaged by fluorescence microscopy. Among 81 men with prostate cancer, 60 men were positive for PPIX (sensitivity 74.1%), and among 57 men with negative biopsy results, 40 were negative for PPIX (specificity 70%).
Proteins in urine
Many proteins have been reported as candidate biomarkers by comprehensive analysis of urinary proteins using array or proteomics technology, but no protein biomarkers have entered clinical use yet. The iTRAQ method or the tandem mass tag methods can label each protein with an isobaric tag that enables the comprehensive quantitative comparison of each protein in samples.8 LC-MS/MS analysis with iTRAQ methods was carried out in urine obtained from men with prostate cancer and BPH without any manipulation of the prostate. Proteins were obtained by organic precipitation with methanol, and six proteins – β2M, PGA3, intestinal mucin (MUC3), apolipoprotein D, alpha-2-glycoprotein 1, ZAG and uromodulin (THP) – were identified by iTRAQ-LC-MS/MS methods as being differentially expressed in men with prostate cancer.9 This group validated these six proteins with immunoblotting, and found β2M, PGA3 and MUC3 to be elevated in urine from the men with prostate cancer. The AUC values of β2M, PGA3 and MUC3 for the detection of prostate cancer were 0.668, 0.625 and 0.618, respectively.
Endoglin (CD105), a type I homodimeric integral transmembrane glycoprotein, was found to be highly expressed in prostatic fluids collected from the radical prostatectomy specimens by cytokine-antibody microarray analysis.10 Endoglin was found to be elevated in urine after massage in prostate cancer patients compared with biopsy-negative men.11 Other proteins, including AMACR,12 IL-18BPa,13 annexin A3,14 engrailed-215 and ZAG16 were also reported to be biomarkers of prostate cancer.
Glycoproteins could be a promising area of research in the development of new biomarkers. Prostate cancer changes the expression of enzymes catalyzing glycosylation, which results in the characteristic glycoproteins seen with prostate cancer. Glycoproteins in the supernatant of urine after prostate massage were blotted by anti-GCNT1 monoclonal antibody, and positive GCNT1 was significantly associated with extracapsular extension of prostate cancer in prostatectomy specimens (AUC 0.76).17 Glycosylation profile (non-fucosylated bi-, tri- and tetra-antennary glycan structures on total of triantennary glycan structures) in urinary protein was found to discriminate BPH (n = 93) from prostate cancer (n = 54). In the gray zone, the AUC of glycosylation profile was 0.81 compared with 0.57 for PSA.18 PSA is also a glycoprotein with one N-glycosylation site (Asn). Serum PSA with aberrant glycosylation was found to be superior to serum PSA itself.19 High levels of PSA were secreted into the prostatic fluids, and a change in glycosylated PSA was also found in urine. Fucosylated PSA levels in urine after massage were measured by lectin-antibody enzyme-linked immunosorbent assay in 69 men (20 men with negative biopsy and 49 men with prostate cancer) and were significantly decreased in the men with prostate cancer compared with the men whose biopsies were negative for cancer. Urinary fucosylated PSA was also associated with the Gleason score. The AUC for the prediction of cancers with a Gleason score ≥7 was 0.72.20
DNA in urine
DNAs originating from prostate cancer are present in urine from men with prostate cancer. Urine precipitates contain prostate cancer cells or fragmented cells that contain the cancer DNAs. The methylation status of the CG island of the glutathione GSTP1 was analyzed in prostatectomy tissue specimens, and the GSTP1 CG island was methylated in prostate cancer specimens.21 In urine precipitates obtained after prostatic massage, GSTP1 promoter hypermethylation was found in 2% of 45 patients diagnosed as having BPH and in 72% of 40 patients with prostate cancer.22 The sensitivity and specificity for the detection of prostate cancer were 73% and 98%, respectively. The significance of the hypermethylation of the GSTP1 promoter in urine precipitates for the detection of prostate cancer was reported by several groups.23-25 RASSF1 promoter methylation was analyzed in urine precipitates from 253 patients with prostate cancer and 32 with BPH, and RASSF1 hypermethylation was found in 45% of the prostate cancer urine samples. The specificity of RASSF1 for cancer detection was 84.4%.26 Methylation of APC, EDNRB, RAR beta2 and HIST1H4K in urinary precipitates was also useful for the detection of prostate cancer.27-30 Recently, a four-gene methylation classifier panel (APC, CRIP3, GSTP1 and HOXD8) predicted patient reclassification (OR 2.559, 95% CI 1.25–5.21) in 153 men under active surveillance.31
DNA from prostate cancer also exists in urine as cell-free DNA. Cell-free DNA was isolated from urine without manipulation of the prostate from patients with CRPC, and AR amplification was detected in five of 10 patients with CRPC. TMPRSS2-ERG fusion, PTEN gene deletion, NOTCH1 locus amplification and MYCL amplification were also found in urine cell-free DNA from CRPC patients.32 However, analysis of urine cell-free DNA by sequencing c-MYC, HER2 and AR was not useful for the detection of prostate cancer in a cohort of 55 patients with prostate cancer (median PSA level of 6.3 ng/mL) and 55 patients with benign disease (AUC 0.504) compared with serum PSA (AUC 0.842).33
RNA in urine
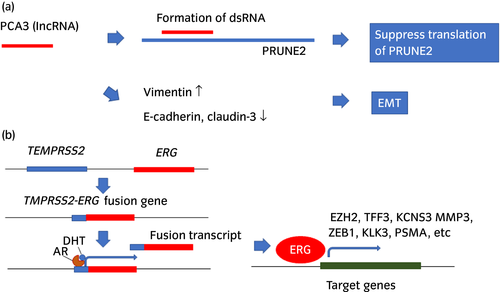
PCA3 is the most widely used urinary marker of prostate cancer. PCA3 was originally found as DD3 in 1999 by Bussemakers et al. They used differential display analysis to compare the mRNA expression pattern of normal versus malignant prostate tissues and identified a cDNA, DD3, which was highly overexpressed in 53 of 56 prostate cancer specimens.34 Real-time PCR methods were developed for the quantification of DD3 (PCA3),35 and DD3 (PCA3) was found to be detectable in urine precipitates after prostate massage. Of 24 men with positive biopsy results, 16 were positive for DD3 (PCA3), and the sensitivity and negative predictive values were 67% and 90%, respectively.36 Subsequently, DD3 (PCA3) was found to be a long non-coding RNA, and overexpression of DD3 (PCA3) induced downregulation of PRUNE2, which harbors the DD3 (PCA3) locus, leading to cell proliferation (Fig. 2a).37 PCA3 knockdown induced the upregulation of several transcripts coding for AR cofactors and modulated the expression of EMT markers. Knockdown of PCA3 resulted in the upregulation of E-cadherin, claudin-3 and cytokeratin-18, and the downregulation of vimentin.38 The PROGENSA PCA3 urine test measures the PCA3 RNA and PSA mRNA used for the normalization. The PCA3 urine test was Food and Drug Administration-approved in the USA in 2012 for men with a previous negative biopsy.39 PCA3 tests were carried out in 1140 men who entered the placebo arm of the REduction by DUtasteride of prostate Cancer Events trial. These men had a negative prostate biopsy at baseline and moderately increased serum PSA levels. PCA3 scores were significantly associated with a positive re-biopsy rate and correlated with the biopsy Gleason score.40 The AUC of the optimal model with PCA3 for cancer detection was 0.75 compared with an AUC of 0.71 for the optimal model without PCA3. A meta-analysis of 46 clinical trials including 12 265 men who underwent PCA3 testing showed that the sensitivity and specificity for the detection of prostate cancer were 65% and 73%, respectively, although this meta-analysis did not separate men with a first biopsy and a repeat biopsy.41 A PCA3-based nomogram for the prediction of prostate cancer and high-grade prostate cancer at the time of initial biopsy was also developed using the information of age, serum PSA, DRE prostate volume and PCA3 level.42, 43 Inclusion of PCA3 into the nomogram increased the accuracy of prostate cancer detection by 4.5–7.1%. The PCA3 scores obtained at the first biopsy and during active surveillance were significantly higher in the patients with Gleason grade reclassification than in those without this reclassification, but the longitudinal change in the PCA3 score during active surveillance did not differ according to the status of Gleason grade reclassification.44
ERG
Gene fusions between TMPRSS2 and ETS in prostate cancer cells were first discovered in 2005.45 TMPRSS2 is a prostate-specific androgen responsive protease. The most frequent gene in the ETS gene family fusing with TMPRSS2 was the ERG gene (present in approximately 85% of all ETS fusion-positive samples). TMPRSS2 has 14 exons and ERG has 11 exons. Several types of fusion genes were reported, and the most common type is the fusion between TMPRSS exon 1 and ERG exon 4–11. Fusion of these genes resulted in the upregulation of ERG. Overexpressed ERG can induce its target gene expression, such as EZH2, TFF3, KCNS3, MMP3, ZEB1, KLK3 and PSMA (Fig. 2b). The prevalence of TMPRSS2-ERG was 30–50% in patients with localized prostate cancer. TMPRSS2-ERG was also detected in urine after DRE from men with prostate cancer.46 Eight of 19 patients with prostate cancer (43%) had detectable TMPRSS2-ERG transcripts in their urine.
A prospective study of 1077 men who underwent their first prostate biopsy was carried out to evaluate the combined measurement of PCA3 and TMPRSS2-ERG RNA in the urine after prostate massage.47 Among 516 men in the developmental cohort, the sensitivity for detecting aggressive prostate cancer was 95%, and the specificity was 39% by combining PCA3 and TMPRSS2-ERG. Among the 561 men in the validation cohort, the sensitivity was still 93% and the specificity was 33% for the detection of aggressive prostate cancer. By combining PCA3 and TMPRSS2-ERG, 42% of unnecessary biopsies could be averted. Recently, it was reported that the benefit of PCA3 and TMPRRS2-ERG differed among races.48 African American men did not show a benefit with the use of PCA3 or TMPRRS2-ERG for the detection of prostate cancer and clinically significant cancer, whereas non-African American men did show benefits. The clinical utility of new biomarkers should thus be tested in each race.
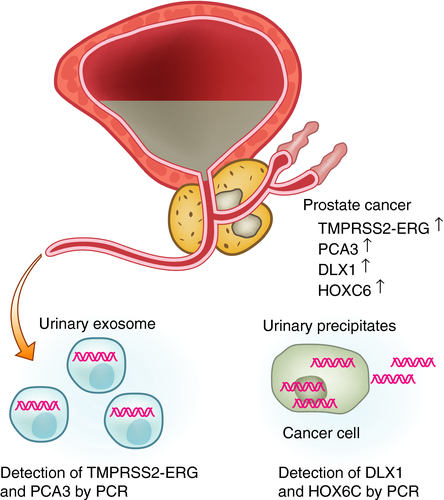
As the values of the PROGENSA PCA3 test and TMPRSS2-ERG for the detection of aggressive prostate cancer are controversial, researchers are continuing to search for new biomarkers of aggressive prostate cancer. Microarray analysis of mRNA from prostate cancer tissues compared with normal prostate revealed 39 potential biomarker candidates.49 Among them, eight mRNAs (HOXC4, HOXC6, DLX1, TDRD1, ONECUT2, NKAIN1, MS4A8B and PPFIA2) were upregulated in precipitates of urine obtained after DRE from patients with prostate cancer compared with men without prostate cancer. Furthermore, a three-gene panel (HOXC6, TDRD1 and DLX16) showed higher accuracy (AUC 0.77) to predict prostate cancer of Gleason score 7 in biopsies compared with the PROGENSA PCA3 test (AUC 0.68) or serum PSA (AUC 0.72). These mRNAs (PCA3, TDRD1, DLX1, HOXC4 and HOXC6) were quantified by using one-step reverse transcription quantitative polymerase chain reaction in 905 urine samples. The KLK3 gene, encoding for PSA, was used as a reference for relative biomarker quantitation. HOXC6 and DLX1 mRNA levels were shown to be good predictors for the detection of high-grade prostate cancer (Gleason score ≥7; AUC 0.76).50 HOXC6 is a developmental transcriptional factor, and the suppression of HOXC6 resulted in decreased cell viability and induction of apoptosis through neutral endopeptidase and IGFBP-3 in prostate cancer cell lines.51 DLX1 is a binding protein of beta-catenin, and promotes the growth, migration and colony formation of prostate cancer cells by activating beta-catenin/T-cell factor signaling.52 Currently, SelectMDx is available for the detection of prostate cancer. It measures mRNA levels of DLX1 and HOXC6 in urine after prostate massage, and uses KLK3 expression as the internal control (Fig. 3). The SelectMDx score was also associated with multiparametric magnetic resonance imaging outcomes and outperformed the PCA3 test.53
MiR in urine
MiRs are short non-coding RNAs with a length of 18–25 nucleotides that can prevent protein expression through the breakdown of specific target mRNAs or through the inhibition of their translation.54 MiRs can be detected in urine as a free form, in prostate cancer cells in urinary precipitates and in exosomes. MiR-107 and miR-574-3p were significantly higher in urinary precipitates from 70 patients with local prostate cancer and 48 patients with advanced prostate cancer compared with 17 controls. The AUC of miR-107 for the detection of prostate cancer was 0.74, that of miR-574-3p was 0.66 and that of PCA3 was 0.61.55 MiR-205 and miR-214 were also significantly decreased in urine from 32 patients with prostate cancer compared with 12 healthy controls. These microRNAs were chosen from the microarray analysis of prostatectomy specimens. The sensitivity and specificity by the combination of two microRNAs were 89% and 80%, respectively.56 Microarray analysis of microRNAs in urine showed that miR-1825 and miR-484 were significantly elevated in urine from eight prostate cancer patients compared with 22 men without prostate cancer.57 The respective sensitivities and specificities for the detection of prostate cancer were 60% and 69% by miR-1825, and 80% and 19% by miR-484. Mir-183 and miR-205, which were elevated in the prostate cancer specimens, were measured in the precipitates of 76 urine samples after DRE, but the sensitivity and specificity for the detection of prostate cancer were 90% and just 3%, respectively.58 Thus, not all microRNAs elevated in prostate cancer specimens can be used as urinary markers of prostate cancer.
EVs (exosomes) in urine
EVs are small vesicles of 30–1000 nm in diameter that are secreted from various cell types, normal epithelial cells, immune cells and cancer cells. EVs in urine after prostate massage include exosomes and prostasomes. Exosomes are 30–200 nm in diameter, and prostasomes are 40–500 nm in diameter.59 It is not clear whether these vesicles with different sizes have different functions. EVs have been known for many years and were believed to be waste from cells, into which cellular “garbage” was loaded.60 However, recent reports have shown that cells use EVs for cell–cell communication with proteins, mRNAs and microRNAs loaded in them. EVs are found in several bodily fluids, including serum, urine, saliva, cerebrospinal fluids and ascites. They are secreted from cells through the mechanisms of exocytosis and endocytosis, but the details of these mechanisms are still unknown. In 2009, Nilsson et al. reported that prostate cancer-derived exosomes, which contain the prostate cancer-specific markers of PCA3 and TMPRSS-ERG, were detected in urine after prostate massage of patients with prostate cancer.61 Since then, several reports have been published on urinary biomarkers of exosomes (Table 2).
| Urine | Sensitivity | Specificity | AUC | Reference | |
|---|---|---|---|---|---|
| RNAs | |||||
| PCA3, ERG and SPDEF | No manipulation | 0.73 | 59 | ||
| Protein | |||||
|
TMEM256 ADIRF PCYOX1 |
No manipulation, morning urine | 0.87 | 63 | ||
| FABP5 | After massage | 0.75 | 64 | ||
| MicroRNAs | |||||
| miR-145-5p | No manipulation, morning urine | 0.62 (serum PSA: 0.80) | 65 | ||
|
miR-196a-5p miR-501-3p |
After massage |
0.73 0.69 |
66 | ||
| miR-21 and miR-375 | No manipulation | 0.87 | 67 | ||
|
miR-19b miR-125b |
No manipulation |
79% 86% |
95% 65% |
68 | |
| Metabolites | |||||
| Combination of 27 metabolites | No manipulation, morning urine | 88% | 92% | 69 | |
The ExoDx Prostate test is commercially available as a urinary test for the detection of prostate cancer. This urinary exosome gene expression assay measures the three genes, PCA3, ERG (including the TMPRSS2-ERG fusion gene) and SPDEF (sterile alpha motif-pointed domain-containing Ets transcription factor) by reverse transcription quantitative polymerase chain reaction (Fig. 3). SPDEF RNA levels were used for the normalization of PCA3 and ERG RNA levels. First-catch urine samples without prostate manipulation were collected, and exosomes were isolated by the EXOPRO Urine Clinical Sample Concentrator Kit. In the validation cohort of 519 men, the AUC of the urinary exosome gene expression assay with PSA was 0.73 compared with an AUC of PSA of 0.63.62 mRNA levels of PCA3, ERG and KLK3 were compared in whole urine, urinary precipitates (cell pellets) and exosomes, and the influence of DRE was also evaluated.63 All three mRNAs were increased in the urine after DRE compared with the urine without DRE. These mRNA levels were the highest in whole urine, second highest in the exosomes and lowest in the cell pellets. Thus, the sensitivity of the ExoDx Prostate test can be increased if urine obtained after DRE is used.
Even if the PCA3 or ERG levels in EVs were lower than those in whole urine, the analysis of EVs would be worthwhile, especially in proteomic analysis. Whole urine contains high backgrounds of secreted proteins, such as THP. THP, the most abundant protein in urine, is secreted from the thick ascending limb of the loop of Henle.64, 65 THP can interfere with the detection of minute levels of biomarker proteins, and it is important to remove THP before proteomics analysis. Isolation of exosomes can result in the removal of THP from the samples. EVs isolated from morning urine without massage were analyzed by mass spectrometric analysis and quantified by label-free quantification (intensity-based absolute quantification).66 TMEM256, ADIRF and PCYOX1 were significantly elevated in urinary exosomes from patients with prostate cancer compared with healthy volunteers. Proteomes of EVs isolated from urine after massage by ultracentrifugation were also comprehensively analyzed by iTRAQ-labeling methods and selected reaction monitoring/multiple reaction monitoring analysis, and FABP5 was found to be elevated in men with prostate cancer compared with men with negative biopsy, and also to be associated with the Gleason score.67 Granulin, AMBP, CHMP4A and CHMP4C were also elevated in men with high Gleason score prostate cancer.
MicroRNAs in urinary exosomes are also reported to be biomarkers for prostate cancer. Exosomes were isolated in morning urine collected from 60 prostate cancer patients, 37 BPH patients and 24 healthy men, and miR-145-5p, miR-141-5p, miR-1290 and miR-572 were measured by PCR. All microRNAs were significantly elevated in the prostate cancer patients, and miR-145-5p was significantly associated with the Gleason score.68 However, the AUC of miR-145-5p for the detection of prostate cancer was 0.62, worse than that of serum PSA (0.80). Sequencing of microRNAs in exosomes isolated from urine of men with or without prostate cancer revealed that microRNAs (miR-196a-5p, miR-34a-5p, miR-143-3p, miR-501-3p and miR-92a-1-5p) were significantly decreased in prostate cancer patients compared with healthy men. In an independent cohort of 28 prostate cancer patients and 19 healthy men, miR-196a-5p and miR-501-3p were significantly decreased in the urine from the prostate cancer patients by qPCR, and the AUCs of miR-196a-5p and miR-501-3p were 0.73 and 0.69, respectively, for the detection of cancer.69 Other groups reported that miR-21 and miR-375 in urinary exosomes were elevated, but that miR-19b and miR-125b were decreased in prostate cancer patients.70, 71
Exosomes can contain the characteristics of the metabolites in prostate cancer cells. The metabolome analysis of urinary exosomes from 62 patients with prostate cancer and 42 healthy individuals showed significant differences in 28 metabolites between the exosomes from patients with prostate cancer and those from healthy individuals, of which 27 metabolites were significantly lower in the exosomes from the men with prostate cancer.72 The sensitivity and specificity for the detection of prostate cancer by combining these metabolites were 88% and 92%, respectively, although validation studies in independent cohorts were necessary. Other groups also reported that the levels of glucuronate, d-ribose 5-phosphate and isobutyryl-l-carnitine were significantly lower in urinary exosomes from three patients with prostate cancer compared with three healthy men aged <35 years.73
Volatile small molecules
Taverna et al. reported that a well-trained canine could identify the urine from patients with prostate cancer through its olfactory system.74 Urine samples from 362 prostate cancer patients and 540 healthy men were smelled by two German Shepherd explosives detection dogs, and the sensitivity was 98.6–100% and specificity was 97.6–98.7%. This report suggested that some volatile compounds specific to prostate cancer patients likely exist in urine. Volatile organic compounds in urine without prostate massage were investigated by gas chromatography/mass spectrometry.75 A total of 59 urine samples from patients with prostate cancer and 43 urine samples with negative biopsy results were analyzed, and the combination of four volatile organic compounds (detection accuracy 63–65%), 2,6-dimethyl-7-octen-2-ol, pentanal, 3-octanone and 2-octanone, could be used to detect prostate cancer marginally better than serum PSA (detection accuracy 62–64%). The advent of new technology could open up new detection methods based on olfactory systems.
Prognostic markers
Only a few urinary markers were reported to be associated with prognosis (Table 3). Prostatic acid phosphatase and galectin-3 were significantly decreased in urine without manipulations from patients with biochemical relapse after prostatectomy (n = 16) compared with patients without relapse (n = 52).76 PCA3 test at the first follow-up biopsy was a significant predictor of upgrading to Gleason score ≥7 among 90 men with 5α-reductase inhibitor during active surveillance. PCA3 test before prostatectomy was associated with tumor volume and extracapsular extension (n = 72).77 PCA3 test collected from 42 men before biopsy and 118 men on the day before radical prostatectomy was also associated with high Gleason score, pathological stage (≥pT3), tumor volume and positive surgical margins in prostatectomy specimens.78 However, there were no reports showing the direct association of PCA3 with cancer-specific survival or biochemical-free survival.
| Prognosis | Reference | |
|---|---|---|
| Protein | ||
| Prostatic acid phosphatase, galectin-3 | Different expression between patients with or without biochemical relapse after prostatectomy | 75 |
| RNA | ||
| PCA3 | Predict the upgrading to Gleason score ≥7 during active surveillance | 76 |
| PCA3 | Predict the tumor volume, extracapsular extension and positive surgical margins in prostatectomy specimens | 77, 78 |
Conclusions
Urine, which can be obtained non-invasively, contains many biomarkers derived from prostate cancer, and it appears to be a promising source for the discovery of new biomarkers of prostate cancer, although several hurdles still exist. Biomarkers in urine can be affected by urinary volume or other substances existing in urine. To date, four urinary tests for prostate cancer are commercially available (Table 4). Further innovation of techniques to quantify minute substances is expected to result in the ground-breaking discovery of new biomarkers of prostate cancer.
| Urinary biomarkers | Type of tests | Target molecules | Methods |
|---|---|---|---|
| Progensa PCA3 | lncRNA in urine after massage | PCA3 | PCR |
| ExoDx prostate (IntelliScore) | Exosomal RNA in urine without manipulation | ERG, PCA3 and SPDEF | PCR |
| SelectMDx for prostate cancer | mRNA in urine after massage | HOXC6, DLX1 and KLK3 | PCR |
| Mi-Prostate Score | RNA in urine after massage | TMPRSS2-ERG and PCA3 | PCR |
Conflict of interest
None declared.




