Machine learning applications in the diagnosis of leukemia: Current trends and future directions
Abstract
Machine learning (ML) offers opportunities to advance pathological diagnosis, especially with increasing trends in digitalizing microscopic images. Diagnosing leukemia is time-consuming and challenging in many areas globally and there is a growing trend in utilizing ML techniques for its diagnosis. In this review, we aimed to describe the literature of ML utilization in the diagnosis of the four common types of leukemia: acute lymphocytic leukemia (ALL), chronic lymphocytic leukemia (CLL), acute myeloid leukemia (AML), and chronic myelogenous leukemia (CML). Using a strict selection criterion, utilizing MeSH terminology and Boolean logic, an electronic search of MEDLINE and IEEE Xplore Digital Library was performed. The electronic search was complemented by handsearching of references of related studies and the top results of Google Scholar. The full texts of 58 articles were reviewed, out of which, 22 studies were included. The number of studies discussing ALL, AML, CLL, and CML was 12, 8, 3, and 1, respectively. No studies were prospectively applying algorithms in real-world scenarios. Majority of studies had small and homogenous samples and used supervised learning for classification tasks. 91% of the studies were performed after 2010, and 74% of the included studies applied ML algorithms to microscopic diagnosis of leukemia. The included studies illustrated the need to develop the field of ML research, including the transformation from solely designing algorithms to practically applying them clinically.
1 BACKGROUND
Augmented human intelligence (AHI) and artificial intelligence (AI) tools might shape the future of medical practice. The expansion of data generated by our systems, medical literature, and the inefficiencies of healthcare systems will necessitate utilizing the power of AI tools.1, 2 The integration of AHI tools into medical practice, including machine learning (ML) and deep learning algorithms, has begun. For instance, the United States food and drug administration (US-FDA) has approved many AI-based softwares since 2017 for medical use.2, 3 The introduction of digital pathology has brought many opportunities to the field of pathology, such as telemedicine.4, 5 Recently, the use of digital pathology has allowed for the use of ML (including deep learning algorithms) in the automation of pathological diagnosis.6, 7 The challenges facing the use of ML in pathology are many, including digitalizing slides, labeling in case of supervised learning, initial and maintenance costs, advanced equipment, technical expertise, and ethical considerations. However, the possible opportunities of implementing AHI tools in pathology are numerous.4, 8, 9
Implementation of ML in pathology has expanded in the last few years. Using whole-slide imaging (WSI), Bejnordi and his colleagues10 presented algorithms submitted as part of a challenge competition to use deep learning to detect lymph nodes with breast cancer metastasis. Seven of the 32 proposed algorithms had significantly higher area under the curve (AUC) when compared to that of 11 pathologists with varying experiences. Moreover, the performance of five algorithms was similar to pathologists, when pathologists were not limited by time. This experiment illustrates the potential advantage of using ML and its promise for achieving efficient workflow with high accuracy. Other successful ML examples in the field of pathology included its reported use in lung and brain tumors.11, 12
Leukemia is a major haematological malignancy that confers mortality and morbidity throughout different ages. It was estimated that there were around 350 000 new cases in 2012 worldwide.13 Leukemia diagnosis is challenged by different factors including lack of healthcare access and misclassification due to lack of experienced personnel.14, 15 Thus, leukemia was one of the potential targets of ML utilization. Multiple articles have investigated the different techniques of segmentation and classification of different blood cells, including white blood cells.16-18 Interest in computer-based diagnosing systems has started six decades ago, with early work on blood and cervical smears.16 The introduction of ML algorithms has developed the approach to computer-based diagnosis. Multiple approaches have been already developed to perform different tasks involved in detecting abnormal blood cells.16-18
In this review, we reviewed the literature pertaining the use of ML in acute and chronic leukemia (both lymphoid and myeloid lineages) diagnosis using microscopy and flow cytometry. The aim of this review was to understand the current trends and limitations and propose future research priorities for the use of ML in leukemia diagnosis. We sought to understand the characteristics including study designs, used techniques, and other characteristics of ML literature, especially in the image-based diagnosis of the four most common types of leukemia.
2 MATERIALS AND METHODS
2.1 Data sources and search strategies
A comprehensive search strategy was performed involving all studies that investigated the role of AHI tools, especially deep learning methods, for leukemia diagnosis. The search included only English language, and the databases searched were Ovid MEDLINE (R) In-Process & Other Non-Indexed Citations and Ovid MEDLINE (R) and IEEE Xplore Digital Library. The search strategies used Boolean logic with MeSH terminology including terms of leukemia and its subtypes (eg, “Leukemia” and “Leukemia, Myeloid/”) and terms pertaining to AHI techniques (eg, “Machine Learning” and “Neural Networks (Computer)”). Using Ovid, leukemia and its subtypes, search was mapped to the following subheadings: Analysis (/an), Cytology (/cy), Diagnosis (/di), Diagnostic Imaging (/dg), Pathology (/pa), and Physiopathology (/pp). Terms “leukemia or leukaemia” were used to search IEEE Xplore Digital Library. Additionally, top results of Google Scholar and references of related and included studies/reviews were screened. The search was performed by two authors independently to assure the collection of all related studies.
2.2 Inclusion criteria
Included publications in this review were studies that investigated the utilization of ML techniques in diagnostic modalities for leukemia (limited to AML, ALL, CLL, and CML) using microscopic images or flow cytometry. This study included only primary studies that used patient-level data; thus, technical/methodological and review studies were excluded from this paper. The models/algorithms had to have validation information. Validation could be internal (eg, using cross-validation) or external (eg, using new validation set or prospective validation). Only articles with full texts were included, and abstracts were excluded due to the lack of enough details. Publication year specified to be from January 2000 until January 2019. Only articles in English language were included.
2.3 Data collection and extraction
Data collected from the articles included: the type of studies, year of publication, and type of leukemia studied, testing and validation set characteristics. In studies investigating algorithms for automated microscopic diagnosis for leukemia, information regarding segmentation, feature extraction, and classification algorithms (supervised/unsupervised) was extracted. Conversely, characteristics of classifier(s) used in case of flow cytometric studies were included. In both types of diagnostic approaches, evaluation metrics (eg, sensitivity, specificity, and AUC) were collected for the proposed algorithm (or algorithm with best outcome if more than one used). The data were collected by two independent researchers, and any variance in collected data was discussed between the two researchers.
3 RESULTS
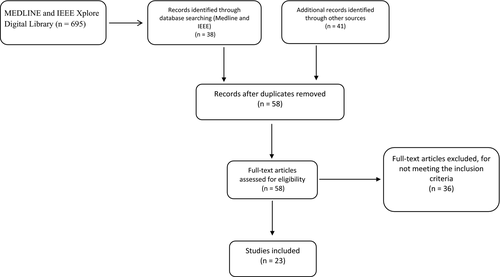
Applying the search strategy in MEDLINE and IEEE Xplore Digital Library, 695 results were initially found. After title and abstract screening, the full texts of 38 studies were reviewed. In addition, handsearching of relevant studies and the top Google Scholar results yielded 40 studies for full-text review. After removing duplicates, twenty-three (23) studies19-41 satisfied the inclusion criteria (see Figure 1). The studies were classified according to the type of leukemia into: ALL (13), AML (8), CLL (3), and CML (1). Two studies proposed diagnostic models for both AML and ALL.
Evaluation metrics used in the studies varied; however, all studies reported at least one metric, with sensitivity and accuracy to be most commonly used. Most of the included studies used supervised learning, with fewer studies using unsupervised learning algorithms for leukemia classification. Most of the studies (21 studies, 91%) were performed after 2010. Majority of the included studies (17 studies, 74%) applied ML algorithms to microscopic diagnosis of leukemia, with 6 included studies (27%) utilized it in flow cytometric diagnosis. None of the studies were prospectively assessing ML models, with k-fold cross-validation method, being the most used to validate the proposed models. Tables 1-3 show the details of the included studies.
| Study | Type of study (Diagnostic modality used) |
Training set Total images (Number of patients) |
Validation strategy Total images (Number of patients) |
Segmentation method | Classifier/s used | Reported evaluation metrics |
|---|---|---|---|---|---|---|
| Rehman et al19 | Retrospective (MS-BM) | NR | 10-fold CV | Threshold-based method | CNN | 97.78% (accuracy) |
| Shafique and Tehsin20 | Retrospective (MS-PBS) | 186 (NR)a | 124 (NR)a | NA | DCNN | ≥94% (sensitivity, specificity, accuracy, and precision)b |
| Rawat et al21 | Retrospective (MS-PBS) | 130 (NR) | 130 (NR) | Threshold-based method | Hybrid hierarchical classifiers (SVM, KNN, ANFIS, PNN) | 99% (overall accuracy) |
| MoradiAmin et al22 | Retrospective (MS-PBS and BM) | 312 (D: 14, O:7) | 10-fold CV | Pattern recognition–based | SVM | >90% (sensitivity, specificity, accuracy, and precision)c |
| Bigorra et al23 | Retrospective (MS-PBS) | 696 (D: 6, O:26) | New cases: 220 images | Pattern recognition–based | SVM | 74% (accuracy) |
| Bhattacharjee and Saini24 | Retrospective (MS-PBS) | 120 (NR) | CV | Pattern recognition–based | Multiple (ANN, kNN, k-means, and SVM) |
100% (sensitivity) 95% (specificity) |
| Rawat et al25 | Retrospective (MS-PBS) | 65 (NR-all ALL) | New cases: 65 images | Threshold-based method | SVM | 87% (accuracy) |
| Reta et al26 | Retrospective (MS-BM) | 633 (D: 34, O:29) | 10-fold CV | Pattern recognition–based | Multiple (KNN, RF, SL, SVM, RC) |
94% (overall accuracy)d 92% ( AUC of AML vs ALL) |
| Chin Neoh et al27 | Retrospective (MS-PBS) | 180 (NR) | 10-fold CV + 500 Bootstrap sampling | Pattern recognition–based | Multiple (MLP, SVM, EC) | 97% (accuracy) |
| Putzue et al28 | Retrospective (MS-PBS) | 30 (NR) | 10-fold CV | Threshold-based method | SVM | 92% (accuracy) |
| Mohapatra et al29 | Retrospective (MS-PBS and BM) | 104 (D: 54, O:50) | 5-fold CV | Pattern recognition–based | Multiple (EC of NB, kNN, MLP, RBFN, SVM, and individually). | 95% (sensitivity, specificity, and accuracy) |
| Ongun et al30 | Retrospective (MS-PBS and BM) | 76 (NR) | 32 (NR) and leave-one-out (LOO) | Deformable models | Multiple (KNN, LV, SVM) | 88% (accuracy) |
| Fiser et al31 | Retrospective (FC) | 123e | 10-fold CV | NA | HCA and SVM | NR |
- Abbreviations: ANFIS, Artificial Neural Network Fuzzy Inference System; ANN, artificial neural network; AUC, area under the curve; BM, bone marrow slides; CV, cross-validation; D, disease; EC, ensemble classifiers; FC, flow cytometry; HCA, hierarchical clustering analysis; KNN, k-nearest neighbor; MLP, multilayer perceptron; MS: microscopic; NB, naive Bayesian; NR, not reported; O, others; PBS, peripheral blood smear; RBFN, radial basis functional network; RC, random committee; RF, random forest; SL, simple logistic; SVM, support vector machine.
- a Data augmentation was used to increase the number.
- b Sensitivity, specificity, accuracy, and precision were ≥94% for L1, L2, L3 ALL subtypes and noncancerous cells. Using multiclass classifiers, the accuracy of diagnosing L1 and L2 was 87% and 86%, respectively.
- c Sensitivity, specificity, accuracy, and precision were above >90 for L1, L2, L3 ALL subtypes and noncancerous cells.
- d Using multiclass classifiers, the accuracy of diagnosing L1 and L2 was 87% and 86%, respectively.
- e Total number of patients.
| Study | Type of study (Diagnostic modality used) |
Training set Total images (Number of patients) |
Validation strategy | Segmentation method | Classifier/s used | Reported evaluation metrics |
|---|---|---|---|---|---|---|
| Bigorra et al23 | Retrospective (MS-PBS) | 696 (D: 11, O:21) | New cases (220 images) | Pattern recognition–based | SVM | 82% (accuracy) |
| Kazemi et al32 | Retrospective (MS-PBS and BM) | 330 (D: 17, O:10) | 10-fold CV | Pattern recognition–based | SVM | ≥95% (sensitivity, specificity, and accuracy) |
| Reta et al26 | Retrospective (MS-BM) | 633 (D: 29, O:34) | 10-fold CV | Pattern recognition–based | Multiple (KNN, RF, LR, SVM, RC) |
97% (overall accuracy)a 92% (AUC of AML vs ALL) |
| Goutam and Sailaja33 | Retrospective (MS-PBS) | 99 (NR) |
CV Hold out LOO |
Pattern recognition–based | SVM | 98% (accuracy) |
| Agaian et al34 | Retrospective (MS-PBS) | 80 (NR) |
CV LOO Hold out |
Pattern recognition–based | SVM | ≥90% (sensitivity, specificity, and precision) |
| Dundar et al35 | Retrospective (FC) | D: 155, O: 0 | D: 43, O:166 | N/A | ASPIRE | 99% (AUC-ROC) |
| Mannien et al36 | Retrospective (FC) | D: 43, O: 316 | 10-fold CV | N/A | SLR and LDA |
100% (accuracy) 98% (AUC-ROC) |
| Biehl et al37 | Retrospective (FC) | D: 23, O:156 | Random selection from the training set | N/A | GMLVQ | 100% (AUC-ROC) |
- Abbreviations: ASPIRE, anomalous sample phenotype identification with random effects; AUC, area under the curve; BM, bone marrow; CV, cross-validation; D, disease; FC, flow cytometry; GMLVQ, Generalized Matrix Relevance Learning Vector Quantization; KNN, k-nearest neighbor; LDA, linear discriminant analysis; LOO, leave-one-out; LR, logistic regression; MS, microscopic; NR, not reported; O, others; PBS, peripheral blood smear; RC, random committee; RF, random forest; SVM, support vector machine.
- a Using multiclass classifiers, the accuracy of diagnosing M1, M2, and M3 was 100%.
| Study | Type of study (Diagnostic modality used) |
Training set No. of images (no. of patients) |
Validation strategy | Segmentation method | Classifier/s used | Reported evaluation metrics |
|---|---|---|---|---|---|---|
| Alferez et al38 | Retrospective (MS-PBS) | 4389 (105) | New cases: 21 patients | Pattern recognition–based | SVM | 91% (overall accuracy) |
| Alferez et al39 | Retrospective (MS-PBS) | 1500 (NR) | 10-CV + New cases: 150 images | Pattern recognition–based | LDA | 80% (accuracy) |
| Lakoumentas et al40 | Retrospective (FC) | NR | New cases: 30 patients | NA | Multiple (BC, FCM, K-means and medians, and SVM) | 99.6% (accuracy) |
- Abbreviations: BC, Bayesian clustering; BM, bone marrow; CV, cross-validation; D, disease; FC, flow cytometry; FCM, fuzzy c-means; LDA, linear discriminant analysis; MS, microscopic; NR, not reported; O, others; PBS, peripheral blood smear; SVM, support vector machine.
3.1 Acute lymphoid leukemia (ALL)
Compared to other leukemia subsets, pathology diagnosis of ALL was the subset with the higher number of studies. Of the included studies, 13 studies investigated the role of ML tools in ALL diagnosis, with 12 studies applied ML tools on microscopic diagnosis and one study applied them on flow cytometric diagnosis.19-31 Seven of the included studies, applying ML on microscopic diagnosis, used only peripheral blood smears, with four studies using bone marrow slides along with blood smears. Only one study solely used bone marrow slides. Refer to Table 1 for further details. None of the studies included were prospectively applying ML models to patients' care, and all were dependent on retrospective data and compared the model performance to a previous diagnosis of cases.
The sample size of blood smears/bone marrow slides varied (whenever reported) between 6 and 120 patients. However, all studies used multiple images from the same patient as well. For instance, Bigorra et al23 utilized a total of 696 images from ALL and non-ALL patients to develop an algorithm. As previously mentioned, none of the proposed algorithms were validated prospectively in clinical settings; however, a variety of validation methodologies were used including training sets or independent preset validation sets. Eight studies have utilized cross-validation as a validation technique for their models. Chin Neoh et al27 used bootstrap sampling in addition to cross-validation. On the other hand, Shafique & Tehsin et al20 used new set of images to serve as a validation technique.
Studies pertaining microscopic diagnosis of ALL commonly followed this sequence: image preprocessing, segmentation, feature extraction, classification, and validation (refer to Figure 2). Image acquisition in the included studies was using either web-based digital libraries or local image repositories. A widely used digital library was ALL-Image DataBase (IDB), which was used in 5 studies (42%).42 ALL-IDB has two data sets, data set (1) cells are not segmented thus allowing for both segmentation and classification exercises, whereas data set (2) cells are segmented.

Included studies followed different approaches for segmentation of ALL cells. The most common segmentation algorithm methodology was pattern recognition–based (eg, fuzzy c-mean and k-means), followed by threshold-based methodologies (eg, watershed). Only one study used deformable models (snakes) as a segmentation methodology. Most of the studies used to segment both nucleus and cytoplasm, with fewer studies that used only nucleus segmentation. No apparent difference in evaluation metrics was noted between studies that segmented only nucleus and the ones that segmented both nucleus and cytoplasm. Feature extraction can be geometric or texture (eg, first and second statistical features). This was not included in Table 1 as all studies used both methods of feature extraction.
Most of the algorithms utilized in the included studies were supervised. The use of deep learning and neural networks was limited in the studies included but highly effective. Shafique & Tehsin et al20 used deep convoluted neural networks on 196 images, with using data augmentation to increase the number of images. The model was able to diagnose leukemia and to differentiate between French-American-British (FAB) classifications: L1, L2, and L3. The overall accuracy of the system was 99.5%. The use of unsupervised algorithms has increased over the past few years and it holds opportunities for pathological diagnosis due to the decreased need of labeling and segmentation.
Evaluation metrics used were sensitivity, specificity, accuracy, precision, and rarely AUC. All included studies reported at least one evaluation metric. Most of the studies included used more than one algorithm with support vector machine (SVM) being the most used. Accuracy of algorithms used ranged from 74% to 99.5%. Bigorra et al23 reported an accuracy of ALL detection of 74% using SVM algorithm. Area under the curves are very widely used method of models' evaluation in AI and ML studies; however, only Reta et al26 reported AUC of SVM algorithm to differentiate between ALL and AML.
3.2 Acute myeloid leukemia (AML)
Eight of the included studies investigated the role of ML tools in AML diagnosis, 5 studies developed models for microscopic diagnosis, and 3 studies developed models for flow cytometric diagnosis.23, 26, 32-37 Three studies used peripheral blood smears, one study used bone marrow slides and peripheral blood smears, and one study used bone marrow slides only. Table 2 shows the details of the included studies. As in the case of ALL, none of the studies included were prospective.
The sample size of range (whenever reported) was 11-155 patients, with using multiple pictures from the same patient. For instance, Bigorra et al23 utilized a total of 696 images from AML and non-AML patients (including ALL and normal) to develop the algorithm. Five studies have utilized cross-validation as a validation technique for their models, with Agaian et al and Goutam & Sailaja33, 34 using leave-one-out (LOO) and hold out along with cross-validation. Biehl et al37 randomly selected 75% of the training set to serve as a validation set. Dundar et al and Bigorra et al used independent set to evaluate and validate their models.23, 35
The five included studies pertaining microscopic diagnosis of AML followed different approaches of segmentation; however, all the reported methodologies were based on pattern recognition. Agaian et al34 used k-means clustering to segment the nucleus, whereas Bigorra et al23 used fuzzy c-mean to segment nucleus, cytoplasm, and peripheral zone around AML cells. On the other hand, Reta et al26 used an innovative approach taking into account color and textural characteristics. Similar to ALL, majority of the used classification algorithms were supervised.
Evaluation metrics used were sensitivity, specificity, accuracy, precision, and AUC. All included studies reported at least one evaluation metric, with all flow cytometric studies reporting AUC. Support vector machine was the most used. Accuracy of algorithms used has ranged from 82% to 97% for studies involving microscopic evidence. The range of AUC was 98%-100% for studies investigating flow cytometric diagnosis. Reta et al26 was the only study which developed an algorithm to differentiate between the different subtypes of AML, with reported accuracy of 100% for M2, M3, and M5.
3.3 Chronic lymphoid leukemia (CLL)
Three of the studies included investigated the use of ML tools in diagnosing CLL, two studies aimed to utilize ML in CLL microscopic diagnosis, and one study aimed to utilize ML in CLL flow cytometric diagnosis.38-40 All studies developed models using peripheral blood smears. Table 2 shows the details of the included studies. As in the case of ALL and AML, none of the studies prospectively applied their models in patients' care.
Using pattern recognition–based segmentation, Alferez et al38, 39 conducted two studies applying SVM and linear discriminant analysis (LDA) on microscopic diagnosis of CLL. In 2015, Alferez et al39 used 1500 images to develop a model using LDA. The model was validated using cross-validation and new 150 images, and it yielded an accuracy of 80%. In 2016, Alferez et al38 used 4000+ images to develop a model using LDA. The model was validated by samples from 21 patients and achieved an overall accuracy of 91%. On the other hand, Lakoumentas et al40 achieved 99.6% accuracy in flow cytometric diagnosis of CLL after using multiple algorithms, of which Bayesian clustering (BC) was the most accurate.
3.4 Chronic myeloid leukemia (CML)
Out of the four major leukemia subsets, CML has the least literature investigating the use of ML tools in its microscopic and flow cytometric diagnosis. One study was included in our analysis targeting CML flow cytometric diagnosis.41 Ni et al41 used SVM (a supervised algorithm) to create a model that is able to distinguish CML from normal cytometric analysis. The model was built using the data of 9 CML patients and 9 normal flow cytometric analyses. The proposed model was able to achieve sensitivity and specificity of <95%.
4 DISCUSSION
Leukemia is a major haematological malignancy, with high prevalence and incidence.13 Leukemia diagnosis, worldwide, is facing multiple challenges.14, 15 The improvements in our ML techniques have given the opportunity for implementing these techniques in leukemia diagnosis.16, 18 In this review, we sought to investigate the different uses of ML in microscopic and flow cytometric diagnostic tools of leukemia in both myeloid and lymphoid lineages.
This review has yielded multiple studies for each major leukemia type (CML, AML, CLL, and ALL) applying ML techniques on microscopic and flow cytometric diagnosis. In general, studies pertaining microscopic image diagnosis of leukemia were more in number compared to flow cytometric studies. It can be noted that the leukemia subset with the least amount of studies was CML, which can be attributed to the necessity of genetic diagnosis in CML. Multiple abstracts have been presented in hematology-, pathology-, and technology-related conferences. In 2018 American Society of Hematology meeting, Höllein et al43 investigated the role of AI in multiparameter flow cytometry (MFC) for the diagnosis of B-cell lymphomas and leukemias. Using data of 16 384 patients and controls, a model was developed with using neural networks. The results were validated using a 10-fold cross-validation. The system achieved 97% accuracy in determining normal vs abnormal cells; however, the accuracy was 74% in classifying the subsets of the included B-cell lymphomas and leukemias.
Automated microscopic diagnosis studies used variety of segmentation methodologies of both nucleus and cytoplasm. The most used method was pattern recognition–based method, with fuzzy c-means being the most commonly used methodology. Fuzzy c-mean has shown to be more accurate than k-means clustering.20, 22 All studies have extracted both geometric and texture features. Included studies represented many limitations in AI/ML research. These limitations include issues like sample size, generalizability, and prospective analysis.
It was not infrequent for models presented in this paper to achieve high accuracy (commonly >90%). This is a very common result in the field of ML research in pathology and other fields as well. This might be appealing; however, it might raise different issues. Firstly, the presented models in this review are generally based on a small sample size and in many studies data were from a single center, which raises the question of how generalizable ML models are proposed from the included studies on other groups of patients.44 Thus, there is a need for these studies to use more robust databases that will need registries and huge digital libraries with the ability to avoid the limitation of overfitting.44, 45 Digitalizing pathology slides has been slow compared to radiology.8 This is further confounded by the additional challenges implementing high magnification slide digitization for hematopathology.46, 47 Hematology slides are harder to digitize than surgical pathology slides due to limitation related to scan time, file size, and possible need for Z-stacking. The lack of adequately developed digital imaging platforms will remain to be the main limitation for wide adoption. Another concern is that many of the studies included would have two sets of data: diseased and normal. This “binary” approach to medical problems is not realistic and does not reflect the real-life complexity of pathological diagnosis.9, 48, 49
Another major issue in the included studies was the lack of prospective validation of models. This has been noted in the literature of ML and deep learning. For instance, Topol et al2 reported that out of the five studies using deep learning in pathology (with results compared to physicians), one study only was prospective in nature utilizing AHI in breast cancer metastases.50 Thus, there is a need to develop both data quality and quantity to potentiate the powers of ML tools.51 This study supports that the current quality of studies pertaining ML is suboptimal, and there is an imperative need to improve both quality and quantity of data before the prospective application of these models in medical practice.
Majority of the included studies in this review used supervised learning algorithms. A drawback of using supervised learning in pathology is the need to label samples which is time-consuming and might introduce errors. A solution to that would be to use unsupervised learning methodologies, in which the patterns are determined by the data itself.2, 9 In the medical literature, the use of unsupervised models is still uncommon. The increase in the implementation of unsupervised learning and deep neural networks (DNNs) might allow for the ability to use bigger data sets. The use of bigger data sets, in addition to the intrinsic abilities of DNN, will allow for the development of more accurate future systems. Moreover, we found no publications incorporating various diagnostic methodologies together in ML models, including genomics. Few studies have investigated the role of ML in genetic diagnosis of leukemia, this is an expanding field that should be a component, in addition to microscopic and flow cytometric components, in creating a comprehensive (using ML tools) diagnostic systems for leukemia. For instance, Aghamaleki et al52 applied artificial neural networks to genetic diagnosis of CLL and achieved an area under the receiver operating characteristic (ROC) curve of 0.991. Another application of ML and deep learning was identifying 20 proteins with the strongest association to FLT3-ITD mutation of AML.53
Augmented human intelligence in health care as a field has been targeted by data scientists of different backgrounds (eg, medical, engineering, and statistics). Thus, this review is limited by our focus mainly on medical literature and databases. However, the aim of this review was not merely to quantitatively present studies applying AHI/ML on leukemia diagnosis, but to note the trends and to comment on the limitations of our current approaches. AHI/ML holds great potentials to improve our current healthcare status in different fields, including pathology. Digital pathology and AI combination will lead to improved workflow and increased efficiency, and the use of digital pathology has led to different advancements in automation including the classification of acute leukemia.54, 55 It is unlikely that AHI/ML will replace physicians; however, it will assist physicians to improve health care.56 Evidence-based and thoughtful approaches to AI/ML implementation are required to ensure safe and useful integration.
5 CONCLUSION AND FUTURE DIRECTIONS
- Efforts to digitalize pathological slides should continue, and creating larger libraries for multiple diseases and pathologies is needed. These libraries can serve as robust databases that can be used to train and validate future models. In addition to the quantitative increase in sample numbers, libraries can allow for sets to be more diverse and not limited to a specific population.
- AHI/ML research should develop from creating models to implementing models in real-world clinical practice. Thus, there should be a shift in AHI/ML research toward integrating these models in daily clinical care.
- Diagnostic accuracy of AHI/ML models is not the only advantage; the increased clinical care efficiency in cost and workflow is another major advantage. Thus, research agenda for AHI/ML in leukemia should fulfill the aim of providing a better and more efficient health care. AHI/ML holds the promise of vital role in improving health care reach in underserved areas and speeding up the diagnosis of acute conditions (eg, acute promyelocytic leukemia).
CONFLICT OF INTEREST
None of the authors declare any relevant conflicts of interest. SKH has received honorarium from Mallinckrodt Pharmaceuticals.
AUTHOR CONTRIBUTIONS
HS, IM, and SH wrote the first draft of the manuscript. All authors vouch for the accuracy and contents of the manuscript. All authors approved the final version of the draft. Each author in this research group has contributed to this paper: HTS, INM, MES, TO, and SKH conceive the study. HTS, INM, MES, TO, and SKH designed the study. HTS and INM acquired the data and involved in literature search. HTS, INM, and SKH prepared the manuscript. HTS, INM, MES, TO, and SKH edited and reviewed the manuscript.




