The utility of c-Met as a diagnostic tissue biomarker in primary colorectal cancer
Funding information
Research was supported by internal University of Leeds funding.
Abstract
The transmembrane protein, c-Met, is thought to be overexpressed and activated in colorectal cancer (CRC). This study explored its potential as a diagnostic tissue biomarker for CRC in a large human CRC tissue collection obtained from a randomized clinical trial.
Tissue microarrays of matched normal colorectal epithelium and primary cancer were prepared from specimens obtained from 280 patients recruited to the MRC CLASICC trial (ISRCTN 74883561) and interrogated using immunohistochemistry for c-Met expression. The distribution and intensity of immunopositivity was graded using a validated, semi-quantifiable score, and differences in median scores analysed using the Wilcoxon signed-rank test. A receiver operating characteristic (ROC) curve was plotted to measure the diagnostic accuracy of c-Met as a biomarker in CRC.
Epithelial cell membrane expression of c-Met differed significantly between CRC and normal colorectal tissue: median 12.00 (Interquartile range (IQR) 6-15) versus median 6.00 (IQR 2.70-12.00) respectively (P = <.0001). ROC-AUC analysis of c-Met expression yielded a CRC diagnostic probability of 0.66 (95% CI: 0.61 to 0.70; P < .0001). A score of ≥14.50 showed high specificity at 85.32% (95% CI 80.33%-89.45%) but sensitivity of only 30.92% (CI 25.37%-36.90%).
Thus c-Met is consistently overexpressed in human CRC as compared to normal colorectal epithelium tissue. c-Met expression may have a role in diagnosis and prognostication if combined with other biomarkers.
1 INTRODUCTION
Colorectal cancer (CRC) is the fourth most common cancer in the UK, with around 42,000 newly diagnosed cases each year.1 The majority of CRC is associated with mutations in oncogenes, tumour suppressor genes and DNA mismatch repair genes. These mutations initiate and propagate the adenoma-carcinoma sequence towards the development of malignant disease in the majority of cases.2 Patient survival is strongly influenced by tumour stage at the time of initial diagnosis, with 1-year overall survival rates of 98% vs. 40% for American Joint Committee on Cancer stage 1 and stage 4 respectively.3
Biomarkers are routinely used in clinical practice for diagnosis, to guide prognostication and to predict response to treatment. Expression of the oncofoetal antigen, carcinoembryonic antigen (CEA), is assessed as a blood serum biomarker of CRC and is routinely used to guide postoperative surveillance for recurrent disease.4, 5 The absence of activating Kirsten rat sarcoma viral oncogene (KRAS) and neuroblastoma RAS viral oncogene homolog (NRAS) mutations is confirmed prior to commencing anti-EGFR (anti-epidermal growth factor receptor) monoclonal antibody in advanced CRC 6-9 due to the negative predictive effect of these mutation; however, most wild-type patients still show no significant response to these agents. High expression of the ligands of EGFR, epiregulin (EREG) and amphiregulin (AREG) at the RNA level was deemed highly predictive of therapeutic benefit with the anti-EGFR agent panitumumab in KRAS/NRAS wild-type metastatic CRC in the PICCOLO trial (International Standard Randomised Controlled Trial Number (ISRCTN): 93248876)7, 10; however, routine assessment of these or other predictive tissue biomarkers is not routinely performed in CRC.
The receptor tyrosine kinase, mesenchymal-epithelial transition factor (c-Met), is a transmembrane protein and key mediator of many cellular processes. The expression of c-Met receptor is primarily localized to epithelial tissues and is activated by the ligand hepatocyte growth factor (HGF). Under normal conditions, the formation of this receptor-ligand complex is vital for many processes, including embryogenesis, tissue regeneration, disruption of cell-to-cell adhesion, promotion of morphogenesis and differentiation.11-13 The expression of c-Met has been found to be upregulated in many malignancies, including non-small-cell lung cancer,14 hepatocellular carcinoma15 and CRC.16 Increased expression of c-Met protein in cancer can be stimulated by physiological hypoxia. This adaptive response gives malignant cells a survival benefit within hypoxic tumour microenvironments.17 In recent years, there has been increasing interest in c-Met as a diagnostic and prognostic biomarker, with several studies suggesting that over expression is linked to poorer survival in CRC.18
Our study is the first of its kind to investigate the potential for c-Met to serve as a diagnostic biomarker for CRC by evaluating its expression in a tissue obtained from a randomized controlled trial of resected CRC.
2 MATERIALS AND METHODS
2.1 Trial cohort and ethical approval
The Medical Research Council (MRC) CLASICC trial (ISRCTN 74883561)19, 20 recruited patients with potentially curable CRC to either laparoscopic or open resection. Consent was obtained from participating patients for residual samples of normal colorectal epithelium and cancer to be stored for future research. Ethical approval for use of tissue from the MRC CLASICC trial was obtained from the National Research Ethics Service (London-Dulwich Committee), reference 12/LO/1327 in 2012. Formalin fixed paraffin embedded (FFPE) tissue samples from a representative subgroup of 280 patients were incorporated into seven anonymized tissue microarrays (TMA). Three tumour cores and three matched normal mucosa cores (each 0.6mm diameter) from the most representative area of the archived resection block were incorporated into the TMA.5 Control tissue samples of archival placenta, ovary, prostate, kidney, epididymis, liver, gallbladder, stomach, oesophagus, pancreas, small bowel, lung and muscle were incorporated into the TMA margins for comparative and orientation purposes.
2.2 Immunohistochemical detection of c-met protein in tissue samples
Sections were prepared from the TMA blocks onto SuperFrost Plus Adhesion glass slides (Thermo Fisher Scientific, Altrincham, UK) at 5μm thickness. These were de-paraffinized in xylene and rehydrated in decreasing concentration gradients of ethanol. Heat-induced antigen retrieval was performed by placing the slides into citric acid buffer solution, adjusted to pH 6.2, and microwaved for 10 minutes at 900W. Slides were then incubated with an anti-c-Met monoclonal antibody (1:250, Recombinant anti-c-Met antibody [EP1454Y], Abcam PLC, Cambridge, UK) for one hour at room temperature. Following incubation with the primary anti-c-Met antibody, slides were incubated with SignalStain® Boost IHC Detection Reagent (HRP, Rabbit; Cell Signalling Technology Inc, Leiden, Netherlands) for 30 minutes in a humidified chamber. The SignalStain® 3,3'-diaminobenzidine (DAB) Substrate Kit (Cell Signalling Technology Inc, Leiden, The Netherlands) was used, and the chromogen solution applied for 10 minutes for antibody visualization. In between each incubation step, the slides were washed in a dilute solution of tris-buffered saline solution. Slides were counterstained with Mayer's Haematoxylin solution (Sigma-Aldrich Scientific Inc, Gillingham, UK), dehydrated and mounted on glass cover slips.
2.3 Imaging
Whole slide images were produced at x40 magnification with the Leica BioSystems Aperio AT2 whole slide scanner (Leica Microsystems Ltd, Milton Keynes, UK). Slide images were viewed at x40 magnification and assessed using Aperio ImageScope v12.4.0.5043 (Leica BioSystems Pathology Imaging Ltd, Milton Keynes, UK).
2.4 Scoring c-Met expression
The distribution and intensity of c-Met protein expression was assessed by a trained research fellow, using a previously validated scale.5, 21-23 c-Met protein immunopositivity in epithelial cell membranes from each tissue core was scored. The intensity of the chromogenic reaction was quantified with a numerical scale between 0 and 3 (0 = no detectable expression, 1 = mild expression, 2 = moderate expression and 3 = strong expression). The distribution of membranous c-Met immunopositivity was determined by calculating the percentage of tumour or normal epithelial cells deemed positive as a percentage of all of the tumour/normal epithelial cells in the core total area. For tumour cores, non-cancerous tissue was excluded from the analysis. (The distribution was graded 0 = <5%, 1 = 5%-20%, 2 = 21%-40%, 3 = 41%-60%, 4 = 61%-80%, 5 = 81%-100%.) The intensity and distribution scores generated were multiplied to provide a compound c-Met expression score. Cores with insufficient tumour or normal epithelial tissue for assessment were not included. Control TMA slides were prepared to exclude fixation dependent observations. In total, 263 tumour and 253 normal epithelium core triplicates (or pairs if a single core not assessable) were suitable for analysis and 242 matched tumour-normal tissue paired samples (86.43% of pairs) were included in the final analysis.
The scoring criteria were appraised and approved by all authors in a consensus meeting prior to initiating the scoring process. Representative examples of c-Met expression at each distribution score 0 to 5 and intensity score 0 to 3 were approved by the group and available for reference throughout the scoring process. Independent verification was provided by a second research fellow with appropriate experience of IHC scoring beforehand. Ten per cent of the 242 matched tumour-normal paired samples were selected at random and presented for secondary verification. Where discrepancy arose between the reviewers a consensus decision was reached.
2.5 Statistical analysis
Microsoft Excel and Graphpad Prism 7 (GraphPad V7.04 Software, Inc, California, USA) were used for all data handling and statistical analyses. Wilcoxon matched-pairs signed-rank test was used to compare the median rank difference in scores between the matched (tumour and normal tissue) TMA samples. The interquartile range (IQR) and two-tailed P value are reported. A statistical significance threshold of P = <.05 was applied. The Wilcoxon matched-pairs test estimate of the 95% confidence interval (95% C.I) of the median difference bound by the IQR were calculated. This method used binominal probabilities to predict the 95% C.I around the median and is therefore asymmetrical around the median value provided.24 To account for uncertainty in the data caused by missing core sample pairs, a sensitivity analysis assuming the highest c-Met expression (compound score 15) for all missing data fields was performed.
A receiver operating characteristic curve (ROC curve) was plotted to determine the diagnostic accuracy of c-Met protein expression in CRC. The sensitivity of c-Met for the detection of CRC (true positive rate) was plotted as a function of the specificity (true negative rate) and used to calculate varying decision cut-off thresholds. The area under the curve (AUC) was calculated to numerically quantify the diagnostic accuracy of various c-Met expression scores in discriminating normal epithelial from cancerous tissue.
3 RESULTS
3.1 Population demographics of the CLASICC trial participants
The key demographic and clinicopathological characteristics of the 280 patient cohort from the CLASICC trial are shown inTable 1.5, 19, 20 Entire sample triplicates of seventeen tumour (6.07%) and 27 (9.64%) normal epithelial tissue cores were ungradable.
| Characteristic | Number of cases (%)N = 280 |
|---|---|
|
Age range: 33-93 years Mean age: 68.9 years |
|
| Male | 144 (51.4) |
| Female | 136 (48.6) |
| pT stage | |
| 1 | 10 (3.6) |
| 2 | 54 (19.3) |
| 3 | 161 (57.5) |
| 4 | 52 (18.6) |
| Unknown | 3 (1.1) |
Note
- The utility of c-Met as a diagnostic tissue biomarker in primary colorectal cancer.
3.2 c-Met expression
The normality test was applied and confirmed the populations were not sampled from a normal distribution, and a non-parametric test was required: skewness −0.61 and 0.29 and kurtosis −1.1 and −1.3 for tumour and normal tissue respectively (P <.0001).
Variable c-Met immunopositivity was observed across the tissue core population. CRC demonstrated more frequent strong c-Met expression as compared to normal epithelial mucosa. High expression was defined as an overall c-Met expression intensity score of 15 and was found in 30.80% (81/263) of CRC samples as compared to 12.25% (31/253) in normal tissue samples (P <.0001). No detectable c-Met expression with a composite c-Met expression score of 0 was found in 7.90% (20/253) of normal colorectal tissue samples versus 3.42% (9/263) in matched tumours (P <.0001). Representative examples of c-Met expression are shown in Figure 1.
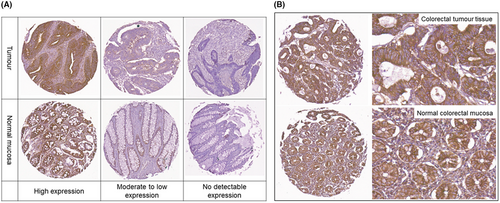
The median tissue c-Met IHC expression scores were 12.0 for tumour (IQR 6.0 to 15.0) and 6.0 for normal epithelium (IQR 2.7 to 12.0) P =<0.0001 (Table 2). Wilcoxon matched-pairs test showed a significant difference between the median expression scores of malignant and normal colorectal tissue, median difference 2.33 (IQR −0.25 to 7.7) and with a difference of sum of signed-rank W score of 15 397 (P <.0001) (n = 242 pairs).
| Tumour | Normal | Difference | |
|---|---|---|---|
| Number of pairs | 242 | ||
| Minimum | 0.00 | 0.00 | −14.00 |
| 25% Percentile | 6.00 | 2.80 | −0.25 |
| Median | 12.00 | 6.00 | 2.33 |
| 75% Percentile | 15.00 | 12.00 | 7.70 |
| Maximum | 15.00 | 15.00 | 15.00 |
| Wilcoxon Matched-pairs Signed-Rank Test | |||
|---|---|---|---|
| P value (two tailed) | <.0001 | <.0001 | |
| 95% C.I of median difference | 1.00 to 3.50 | ||
| IQR median difference | −0.33 to 7.70 | ||
| Sum of signed ranks (W) | 15 397 | ||
| Sum of positive rank | 22 274 | ||
| Sum of negative rank | −6877 | ||
3.3 Accuracy of c-Met as a colorectal cancer diagnostic biomarker
The receiver operating characteristics (ROC) curve and the area under curve (AUC) of the sensitivity and specificity of c-Met expression at various thresholds are shown in Figure 2. The ROC-AUC score yielded an overall diagnostic accuracy predictive probability score of 0.66 (95% CI: 0.61 to 0.70; P = <.0001).
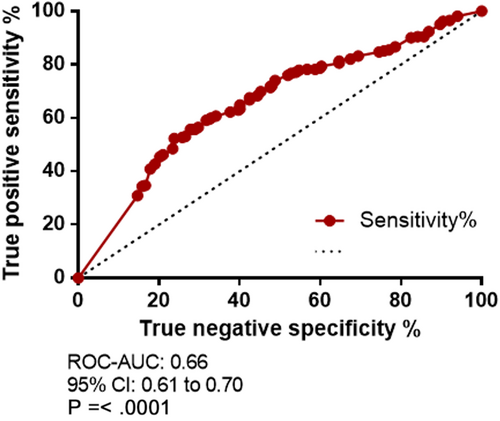
A total expression score of ≥ 14.50 yielded a high specificity of 85.32% (C.I 80.33% to 89.45%); however, there was a significant trade-off with sensitivity of 30.92% (C.I 25.37% to 36.90%) and a likelihood ratio of 2.11. A combined score of the total sensitivity and specificity from each cut-off generated by the ROC curve was calculated. An overall c-Met expression score cut-off of ≥ 11.58, yielded the greatest combined true positive and true negative rate with a sensitivity of 55.73% (95% CI 49.48% to 61.84%) and specificity of 71.83% (95% CI 65.84% to 77.29%), likelihood ratio 1.98. The highest likelihood ratio of 2.29 was observed with a c-Met expression score of ≥ 13.17: sensitivity 40.84% (95% CI 34.83% to 47.06%) and specificity 82.14% (95% CI 76.85% to 86.67%).
3.4 Sensitivity analysis
The sensitivity analysis assumed the highest level of c-Met expression for all missing data (compound score 15). This confirmed a significant difference in median score between the matched tissue samples, 12.30 for tumour (IQR 7.58-15.00; P = <.0001) and 10.00 for normal epithelium (IQR 6.00-13.30; P = <.0001). Similarly, the ROC-AUC analysis showed an AUC value of 0.60 (95% CI 0.55 to 0.64; P = <.0001). However, intense c-Met immunopositivity (expression score ≥14.50) yielded a slightly lower CRC predictive value specificity of 78.47% (95% CI 73.12% to 83.19%) and sensitivity of 33.94% (95% CI 28.35% to 39.88%) with a likelihood ratio 1.19.
4 DISCUSSION
Oncogenic activation of the transmembrane protein, c-Met, has been reported to induce multiple intracellular changes leading to the development and progression of cancers.23, 25-27 Overexpression of c-Met has been reported in several gastrointestinal malignancies, including CRC.24 Previous small studies have identified the overexpression of c-Met in metastatic CRC as a predictor of poor overall and progression-free survival.23, 26, 27 We quantified the expression of c-Met protein in matched tumour and normal epithelial tissue samples from participants of the MRC CLASICC trial. Our study is unique in its utilization of tissue from a multi-centre RCT study cohort to examine a novel biomarker, both in its size and representation of the CRC patient population in the UK.
We adopted a previously validated scoring system5, 21-23 in this study. Tumour samples had a significantly increased median c-Met protein expression, as compared to matched normal mucosa. The c-Met protein plays a vital homeostatic role in epithelial cell function; therefore, the rate of high c-met expression (12.25%) by normal mucosa cores (compared to 30.80% in CRC cores) was entirely expected and observed in previous immunohistochemical studies of c-Met expression.28 Our data support the results of smaller studies; Takeuchi et al 29 confirmed c-Met mRNA copies were significantly elevated in a small sample of CRC patients relative to normal colonic epithelium. Interestingly, their study of 36 patients also identified mRNA copy levels correlated with primary CRC depth of invasion and lymph node status.
Examination of the diagnostic accuracy of c-Met expression in CRC tissue samples confirmed that a total c-Met expression score of 12 or over, yielded the greatest combined specificity and sensitivity for the detection of CRC, at 76.19% and 52.29% respectively. The differential expression and diagnostic accuracy ROC curve assessment indicates that c-Met may have a role in the diagnosis of CRC and could be a possible therapeutic target.
Previous studies have confirmed the potential of tissue CEA as a diagnostic and surveillance biomarker and the CEA receptor protein as an imaging target in CRC.5, 27, 30 Tiernan et al 5 used a similar scoring method to this study and identified increased CEA expression in CRC tissue, when compared to corresponding normal tissue. Although we found that c-Met lacked the diagnostic accuracy of CEA, it outperformed the other biomarkers explored in the Tiernan study: epidermal growth factor receptor (EGFR), Folate receptor alpha (FRα) and tumour-associated glycoprotein 72 (TAG-72). Formation of the proto-oncogene c-Met-HGF ligand complex activates several downstream signalling cascades and induces nuclear transcription and cellular transformation. This can include activation of the RAS protein family.9, 11, 23 KRAS and NRAS mutation status is assessed prior to treatment with anti-EGFR agents in CRC. It is possible c-Met interacts with other known oncogenic pathways in the development of, and chemotherapy resistance seen in CRC Yoon et al confirmed strong correlation between c-Met and a related downstream non-receptor tyrosine kinase, focal adhesion kinase (FAK) expression H-scores in CRC TMA. With the related macrophage-stimulating protein receptor (MST1R), a three-protein risk stratification model of CRC confirmed high FAK, low c-MET and low MST1R protein levels showed the worst progression-free survival and a high risk of early progression of disease. In combination, c-Met may be beneficial in the diagnosis or prognostication of CRC, especially in combination with other novel biomarkers.
5 CONCLUSION
Immunohistochemical analysis of c-Met expression in human tissue samples showed a significant differential expression between matched normal colorectal epithelium and CRC. Malignant tissue consistently overexpressed c-Met protein, relative to normal tissue. Strong c-Met expression only yielded a true negative rate of 30.92%. With such a high false positive rate, the diagnostic and prognostic utility of c-Met in CRC may be limited. Further evaluation of c-Met in conjunction with other biomarkers is required and may have a prognostic, surveillance or response to treatment predictive role.
ACKNOWLEDGEMENT
The CLASICC trial was funded by the Medical Research Council and coordinated by the Clinical Trials Research Unit at the University of Leeds. We thank the patients for their participation and all of the local investigators for delivering the trial. The TMAs were prepared by S Yeluri. M Hale and the Virtual Pathology team within the Division of Pathology and Data Analytics at the Leeds Institute of Medical Research at St. James's assisted with digital slide preparation.
CONFLICT OF INTEREST
None declared.
AUTHORS' CONTRIBUTIONS
G R Armstrong designed and conducted the experiment, analysed the results and drafted the manuscript with guidance from N P West and J P Tiernan and G R Armstrong. J Tiernan previously designed and validated a similar IHC grading classification system and assisted with adapting the method for this project. M I Khot, S L Perry and T I Maisey provided training on laboratory techniques, and T A Hughes, N P West and J P Tiernan assisted G R Armstrong with interpretation of the results. All authors saw and approved the final version of the manuscript.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Tissue was obtained from participants of the MRC CLASICC randomized controlled trial (Medical Research Council Conventional versus Laparoscopic-Assisted Surgery in Colorectal Cancer) (Trial identifiers—ClinicalTrials.gov number NCT00003354 and Clinical Trial Number ISRCTN 74883561). Approval process described previously.24 Ethical approval for tissue research was obtained from the National Research Ethics Service (London-Dulwich Committee), reference 12/LO/1327.
CONSENT FOR PUBLICATION
The authors obtained at the time of CLASICC trial enrolment by local investigators and under National Research Ethics Service (London-Dulwich Committee), reference 12/LO/1327 approval for tissue research.
Open Research
DATA AVAILABILITY STATEMENT
The datasets analysed and results presented in this study are available from the corresponding author on reasonable request.




