Impact of persistently high sea surface temperatures on the rhizobiomes of Zostera marina in a Baltic Sea benthocosms
Abstract
Persistently high marine temperatures are escalating and threating marine biodiversity. The Baltic Sea, warming faster than other seas, is a good model to study the impact of increasing sea surface temperatures. Zostera marina, a key player in the Baltic ecosystem, faces susceptibility to disturbances, especially under chronic high temperatures. Despite the increasing number of studies on the impact of global warming on seagrasses, little attention has been paid to the role of the holobiont. Using an outdoor benthocosm to replicate near-natural conditions, this study explores the repercussions of persistent warming on the microbiome of Z. marina and its implications for holobiont function. Results show that both seasonal warming and chronic warming, impact Z. marina roots and sediment microbiome. Compared with roots, sediments demonstrate higher diversity and stability throughout the study, but temperature effects manifest earlier in both compartments, possibly linked to premature Z. marina die-offs under chronic warming. Shifts in microbial composition, such as an increase in organic matter-degrading and sulfur-related bacteria, accompany chronic warming. A higher ratio of sulfate-reducing bacteria compared to sulfide oxidizers was found in the warming treatment which may result in the collapse of the seagrasses, due to toxic levels of sulfide. Differentiating predicted pathways for warmest temperatures were related to sulfur and nitrogen cycles, suggest an increase of the microbial metabolism, and possible seagrass protection strategies through the production of isoprene. These structural and compositional variations in the associated microbiome offer early insights into the ecological status of seagrasses. Certain taxa/genes/pathways may serve as markers for specific stresses. Monitoring programs should integrate this aspect to identify early indicators of seagrass health. Understanding microbiome changes under stress is crucial for the use of potential probiotic taxa to mitigate climate change effects. Broader-scale examination of seagrass–microorganism interactions is needed to leverage knowledge on host–microbe interactions in seagrasses.
1 INTRODUCTION
Many ecosystems' structure and function are impacted by climate change (Gitay et al., 2002; Schröter et al., 2005). Coastal ecosystems are especially vulnerable because several human activities have already reduced their resilience and ability to withstand new environmental shocks (Holling, 1973; Hughes et al., 2003). Sea surface temperature (SST) trends and regimes are impacted by global warming, resulting in an increase in the mean annual SST. The Baltic Sea is warming faster than most other sea areas in the world (Belkin, 2009; Sherman et al., 2009), representing a warming hotspot (Reusch et al., 2018). As such the Baltic Sea may act as a model for impacts of global warming even though the impacts may be different from those anticipated for oceanic regions. Ecosystem impacts may have both direct and indirect effects on both human society and the biota that live in the Baltic Sea (Hyytiäinen et al., 2019; Paasche et al., 2015; Pihlainen et al., 2020; Stenseth et al., 2020). Seagrasses at large and Zostera marina L. for the Baltic, play a vital role in the ecosystem by providing habitat, primary productivity, sediment stabilization, and nutrient cycling (Den Hartog & Kuo, 2006; Hemminga & Duarte, 2000). Additionally, seagrasses are discussed as one of the planet's most effective carbon sinks and one of the solutions to reduce CO2 emissions and ultimately address the problem of global warming (Gattuso et al., 2018). Because there is little species redundancy, seagrass meadows are particularly susceptible to disturbances like persistently high temperatures (Ehlers et al., 2008), which can inevitably cascade into severe changes in the entire ecosystem. The number of studies on the impact of global warming on seagrasses, as well as their response to this ecological crisis, have been increasing during the last decade (Nguyen et al., 2021).
Despite the increasing number of studies on the impact of global warming on seagrasses (e.g., Ehlers et al., 2008; Hughes & Stachowicz, 2004; Nguyen et al., 2021), little attention has been paid to the impact on host–microbiome interactions, especially in marine ecosystems. This is a crucial gap in our understanding, as bacteria play a vital role in the basal functions of marine plants, such as nutrition, growth promotion, and protection against pathogens. In this context, it is essential to consider the associated microbiomes of plants when evaluating their susceptibility to environmental disruptions. The microbial communities associated with the rhizosphere are considered to be more stable than those associated with the aboveground plant components, which reflect more environmental variability (Conte et al., 2021). So, bacterial community shifts in the plant rhizosphere would give us a better indication of the host physiological changes (García-Martínez et al., 2009).
In terrestrial systems, this approach has already revealed several effects of climate change on the host–microbiome interactions (Cheng et al., 2019). Elevated temperature suppresses several immunity mechanisms in plants, turning them more susceptible to pathogens (Cheng et al., 2013) while increasing virulence of pathogens (Hasegawa et al., 2005). More important in this context, some endophytes and bacteria in the rhizosphere may reduce the detrimental effects of temperature on host plants, increasing their capacity to grow at various temperatures. For example, when tomato roots are inoculated with the strain PsJN of Burkholderia phytofirmans the host tolerance to high temperatures increases (Issa et al., 2018).
Seagrasses harbor a diverse bacterial community associated with their leaves and roots (e.g., Aires et al., 2021; Garcias-Bonet et al., 2016). The crucial role of their bacteria is suggested in some basal functions such as nutrition, growth promotion, and protection against pathogens (see reviews by Conte et al., 2021; Tarquinio et al., 2019; Ugarelli et al., 2017). Compared to the terrestrial ecosystems, dispersal barriers are lower in the marine environment so, we expect more interactions across holobionts allowing greater phylogenetic diversity and easier and faster microbial community shifts (Dittami et al., 2021).
Almost all seagrasses are rooted in soft sediments. Elevated temperatures impact nutrient cycling in sediments (Alsterberg et al., 2011; Currie et al., 2017), due to the way different bacteria respond to those changes. A particular study, examining long-term warming on Baltic coastal sediments, showed an increase of anaerobic processes in the sediment surface and a significant increase in sulfur and nitrogen metabolism-associated genes (Seidel et al., 2022). Shifts in microbiomes of sediments may have a direct and indirect effect on their inhabiting seagrasses and their associated microbiome.
In this study, we investigate the potential impact of ocean warming on seagrass–microbiome interactions in the Baltic Sea, and how this may affect the ecosystem's overall health and function. We argue that studying the role of microbiomes in the response of seagrasses to environmental stress is critical to our understanding of how these ecosystems will respond to future climate change.
In this long-term study, by using an outdoor benthocosms approach, mimicking near-natural conditions (i.e., natural variations in light, temperature, and nutrient availability and including associated organisms as grazers and epiphytes), and applying realistic temperature fluctuations (see Wahl et al., 2015), we aim to investigate the effects of a persistent SST increment on the roots' microbiome and adjacent sediment, of the foundation seagrass species Z. marina. The experiment occurred over a period of 9 months spanning across three seasons (winter–spring–summer—northern hemisphere), with periodic sampling in four time points (early March, mid-April, late May, and late June). In this same experiment, Sawall et al. (2021) registered high mortality of Z. marina under persistent warm treatment after flowering earlier in spring and in ambient seagrasses, after an abnormally warm June. Their main results showed a reduction of 50% biomass in artificially warmed Z. marina suggesting that winter–spring-warmed conditions had severe effects on seagrass vitality (Sawall et al., 2021). In our study, we assess the effect of persistent warming over a 9-month period spanning three different seasons (winter, spring, and summer) on the microbiome associated with macrophytes. This long-term, trans-seasonal study will complement the typically studied summer heatwaves and is crucial for fully understanding the effects of global warming. With this approach, we aim to assess the natural effect of seasonality in both sediments and roots microbiome and for both Ambient (control, natural temperature fluctuation +0°C) and Warm (natural temperature fluctuation +3.6°C) treatments. More importantly, we want to assess the main differences between the bacteria associated with heat-stressed Z. marina individuals and those associated with plants under ambient temperatures. Through bacterial community structure and composition and using functional predictive tools we also aim to understand how bacterial community shift can help seagrasses to cope with chronically increased SST across seasons.
2 MATERIAL AND METHODS
2.1 Experimental design and Z. marina sampling
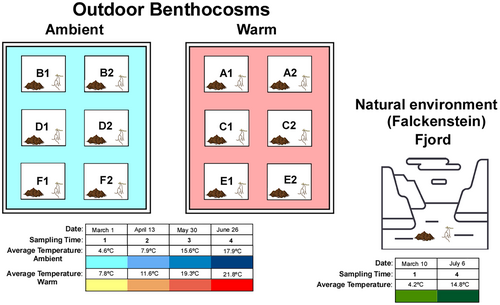
The Kiel Outdoor Benthocosms (KOB), situated on an aluminum float (ALU-BAU, Büdelsdorf, Germany), moored to the dock of GEOMAR Helmholtz Centre for Ocean Research in the inner Kiel Fjord, was used for this experiment from November 25, 2015, to August 26, 2016. The mooring was made up of sledges with wheels that could slide along rails fastened to the pier's supports in order to accommodate for changes in sea level. Six 3000 L benthocosm tanks (Heers & Brockstedt Umwelttechnik, D-24539 Büdelsdorf) with internal dimensions of 2 m × 2 m × 0.9 m and a 5 cm thick styrofoam layer between inner and outer walls, to achieve the best thermal insulation, were housed inside the KOB. These six tanks, each of which divided into two subunits comprising 12 experimental units (1500 L), were labeled A through F and ran in a factorial “ANOVA mode” with present vs. future climatic scenarios (3–6 replicates). Surface seawater from the Kiel Fjord flows through continuously at a rate of 1800 L per day, providing nutrients, organic matter, and daily variations in abiotic parameters like temperature, salinity, pH, and oxygen that are similar to those found in nature. The system includes a wave generator that prevents stratification by causing water movement and it has an open top for natural light supply. A GHL feedback system (GHL Advanced Technology) was specifically installed in each tank, and it worked in tandem with heaters and coolers to maintain the desired temperatures. Wahl et al. (2015) provide complete details on the benthocosms materials, conditions, control, and registering elements (temperature, salinity, oxygen, etc.). The experimental community was created in accordance with the natural shallow-water Baltic Sea system's community structure, which is dominated by Fucus vesiculosus beds and Z. marina meadows. Therefore, the tanks were set up to include 14 common organisms found in such ecosystems, which, among others, included the gastropod Littorina littorea, the isopod Idotea balthica, amphipods of the genus Gammarus, the decapod Palaemon sp., and the bivalve Mytilus edulis. The input of fjord water allowed other organisms to enter the tanks as well. Those were mostly filamentous algae, polychaetes, a few different species of bivalves (including Limecola balthica, Mya sp., and Mysella sp.), and gastropods (like Hydrobia ulvae and Retusa sp.), and the round goby (Neogobius melanostomus). See Sawall et al. (2021) for more details.
Zostera marina was sampled in November 2015 in the Kiel Fjord (Falckenstein: 54°24′24.7″ N, 10°11′38.7″ E), Baltic Sea, along a 100 m transect at 2–3 m depth within a meadow. Around 1–3 shoots (leaves, rhizomes and roots) per ramet were collected and kept in large coolers with fjord water until transported to the KOB within 2 h after collection. Six shoots of similar size (shoot length: 6–10 cm; rhizome length: 3–5 cm), from different plants, were planted, in sediment previously collected from the Fjord, in 5 L boxes. Every tank received 18 of those boxes with six plants each and 10 beakers with two plants each which resulted in a total of 134 plants. During an acclimation period of 7 days, all the shoots were maintained under ambient fjord temperature.
For this particular experiment, two different temperature regimes were applied, with each treatment replicated in six independent tanks: (i) a control regime without simulated temperature rising, but natural stochastic temperature variability (tanks B1, B2, D1, D2, F1, F2 = Ambient); (b) a warming regime, chosen considering a future warming scenario for the Baltic Sea (Gattuso et al., 2014; Gräwe et al., 2013) (tanks A1, A2, C1, C2, E1, E2 = Warm). The temperature regime was set to an increase in around 3.6°C above naturally fluctuating fjord temperatures (Ambient). The target temperature of the Warming treatment was approached by daily temperature increments of 0.3°C over 12 consecutive days, in three different periods. Temperature details for this experiment are represented in figure 1 of Sawall et al. (2021). The number of shoots in each tank was registered a few days before each sampling time.
In order to study, the bacterial community associated with Z. marina roots and adjacent sediment, and compare it between ambient and warming treatments, six replicates (one sample per tank) were collected from the ambient and warming treatments. The sampling was done in four different timings (Mach, April, May, and June) which corresponded to 4 different ambient and warming temperatures (Figure 1). Sediment and plant samples were also collected from the natural Z. marina meadow in the Falckenstein Fjord, in the beginning and end of experiment, to be used as control (Figure 1). All the roots collected for this experiment were taken from living and healthy Z. marina shoots.
Seagrass roots were excised from the plant with sterilized blades and the adjacent sediment was scooped using a 2 mL tube. Samples were stored using the DNA/RNA shield from Zymo Research and stored at −20°C until extraction.
2.2 Abiotic parameters monitoring
Dissolved oxygen was measured continually and logged every 60 s by a Hach-Lange LDO optode in each of the experimental tanks (A, B, C, D, and E) (Hach-Lange, 4H.Jena Engineering GmbH). A GHL Profilux 3ex was used to measure and regulate the temperature, and data were logged every 10 min (the same was done in the Fjord). Around 3 h after daylight, salinity was manually measured using a WTW multimeter (Multi WTW Cond 3110 1 Tetra Con 325). 20 mL of water were taken every month (from December to April) or every 2 months (from April to August) for inorganic nutrient analysis (PO4, NH4, NO2, NO3). This water was filtered using a 0.45 μm cellulose-acetate membrane filter (Sartorius) and the filters were stored at −20°C until further analysis using an autoanalyzer (Seal Analytical) following basic protocols (Lewandowska et al., 2014).
2.3 DNA extraction and bacterial 16S amplicon sequencing
DNA was extracted with the DNeasy PowerSoil Kit (Qiagen, Benelux BV) following the Vortex Adapter protocol. Extracted DNA was sent to Molecular Research (MR DNA, Shallowater, TX, USA). An Illumina library was prepared by amplifying the 16S rRNA gene V3V4 variable region, using the forward 341F (5′-CCTACGGGNGGCWGCAG-3′) and reverse 805R (5′-GACTACHVGGGTATCTAATCC-3′) primers (Klindworth et al., 2013) and sequenced on an Illumina MiSeq system (2 × 250 bp paired-end) following the manufacturer's guidelines. PCR conditions were as follows: 95°C for 3 min, 25 cycles of 95°C for 30 s, 55°C for 30 s, 72°C for 30 s, and a final elongation of 72°C for 5 min. The samples were pooled in equal proportions based on their molecular weight (calculated based on the size of the amplicon) and DNA concentrations (using Qubit Invitrogen) and purified using calibrated Agencourt AMPure XP beads. DNA libraries were prepared by following Illumina MiSeq DNA library preparation protocol and paired-end (2 bp 250 bp) sequencing performed at MR DNA (Shallowater, TX, United States) on an Illumina MiSeq system following the manufacturer's guidelines.
2.4 Bacterial community sequence analysis
Demultiplexed Illumina raw paired-end sequences were imported into the MR DNA software FASTq Processor where barcodes and both primers were removed, and the fastq files were prepared to be used in the Quantitative Insights into Microbial Ecology 2 (Qiime2 v.2021.4; Bolyen et al., 2019) package for complete microbiome analysis. The resulting files were imported into QIIME2 using the Casava 1.8 paired-end demultiplexed fastq tutorial and were then quality filtered, trimmed according to the quality of demultiplexed sequence reads (forward: trimmed left at 0 sequence base and truncated at position 228rd; reverse: trimmed left at 0 sequence base and truncated at position 220th), dereplicated, denoised, merged, and assessed for chimeras to produce ASVs using the DADA2 plugin (Callahan et al., 2016). Taxonomy was assigned to ASVs using a pretrained naive Bayes classifier. The classifier was trained on Silva 138 99% OTUs full-length dataset using the QIIME2 feature classifier plugin (Bokulich et al., 2018). The feature table was then filtered to exclude ASVs assigned to chloroplasts, mitochondria and singletons and doubletons. For statistical purposes, the ASV table was rarefied to the minimum number of sequences (3619). We used rarefied data to perform PERMANOVAs and multivariate analysis, and nonrarefied data to perform any differential/discriminant analyses and make relative abundance bar plots.
The microbial diversity and richness of all samples were estimated using alpha diversity metrics—Shannon index and observed ASVs—using the diversity plugin within QIIME2. Boxplots were generated for a visualization of the alpha diversity differences among groups of samples (grouped according to type of sample, treatment, and sampling time—detailed below). Univariate PERMANOVA was used to determine significant differences (as in Hartmann et al., 2015) among groups of samples. Matrices were constructed using Euclidean distance, and the number of permutations was set to 999. Monte Carlo tests were done when the number of unique permutations was less than 400 (Metropolis & Ulam, 1949).
Bacterial community composition and structure (beta diversity) were assessed through ASV square root transformations, after which a resemblance matrix was compiled using the Bray–Curtis dissimilarity algorithm. Statistical analyses were done using a three-factor PERMANOVA: Type of Sample (fixed factor with two levels—roots and sediment), Temperature treatment (fixed factor with three levels—ambient, warm and control [fjord]), and sampling time (fixed factor with four levels—1, 2, 3, and 4). All the factors and respective interactions were tested. Monte Carlo test was done when the number of unique permutations was less than 400.
Community differentiations were visualized with canonical analysis of principal coordinates (CAP), using the interaction of all three factors (Type of Sample × Treatment × Sampling Time) as a priori factor. For a better visualization of the differences among samples, the control samples (Fjord) were not included in the CAP (but were included in the PERMANOVA statistical tests). Abiotic factors were overlaid as vectors in the CAP plot to show which were the main drivers of the clustering pattern. All the statistical tests and multivariate analysis were performed in PRIMER-E C PERMANOVA v.6 (Clarke & Gorley, 2006).
While using the total bacterial community to produce the relative abundance bar plots, we noticed that some of the most abundant taxa from a given treatment (considering the interaction of the three factors Type of Sample × Temperature Treatment × Sampling Time) were only present in one or a few replicates and were not representative of the complete set of replicates. That would lead us to biased results that would not reflect the consensus community of that certain treatment. Core community usually refers to any group of microbial taxa shared by a certain number of samples from a host species, environment of interest, or treatment (Neu et al., 2021; Risely, 2020). Therefore, we have chosen the core strategy to characterize the bacterial community of each treatment, maintaining only those ASVs that were present in at least 80% of the replicates from that treatment. That way, we would avoid drawing inferences based on abundant but unrepresentative taxa. The core community of each group/treatment of samples (e.g., Roots/Ambient/Sampling time 1) was calculated using the core features function in Qiime 2 (Bolyen et al., 2019) and using 0.80 as the minimum fraction. After each calculation per treatment, all the “core tables” were merged. This “core” dataset was used for bacterial community composition analysis through relative abundance barplots and any comparison and classification analysis (i.e., LEfSe, including for the PiCRUSt analysis).
It is important to highlight that this dataset we called “core” is not the same as the so called “core community” that are constant across seasons, environmental stress, geographic location, etc., for a certain species. Instead, this “core” dataset identifies the main changes from one treatment to another for all or most of the replicates (making it consistent).
Relative abundance barplots were made to visualize the differences in taxa (Family level) between treatments. To assess which taxa strongly discriminate between Ambient and Warm treatments, per sampling point (roots and sediments separately) and to have a clearer idea of the persistently high temperatures effect on the microbiome, LEfSe analysis (Linear discriminant analysis Effect Size) were performed using the online tool Microbiome Analyst (https://www.microbiomeanalyst.ca/; Chong et al., 2020; Dhariwal et al., 2017). LEfSe employs Kruskal–Wallis rank sum test to detect features with significant differential abundance, followed by Linear Discriminant Analysis to evaluate the relevance or effect size of differential abundant features. We set the p-value cutoff to 0.1, using the FDR-adjusted data and 2.0 Log LDA score. Similarly, LEfSe analysis was done to access the main differences between the control/fjord samples and the corresponding (time 1 and 4) ambient sediments and roots.
PERMANOVA analysis and visualization of microbial community structure among distinct treatments (CAP) were done with both datasets (total community and core community) to check discrepancies, and the results were fundamentally the same.
2.5 Predictive functional analysis
Core ASVs table was analyzed using PiCRUSt2 (Phylogenetic Investigation of Communities by Reconstruction of Observed States; Douglas et al., 2020). A cutoff nearest-sequenced taxon index value of 0.15 was employed to remove unreliable predictions, as recommended by Langille et al. (2013). The metabolic functions of these pathways were referenced against the MetaCyc database (Caspi et al., 2018) for description.
The predicted pathway abundances were statistically analyzed, for roots and sediment separately, through a two-factor PERMANOVA: Temperature treatment and sampling time (and their interaction). The parameters used were the same as for the ASV abundance and bacterial community composition (see above). Also, as done for the community composition analysis (see above), CAP plots were prepared to visualize samples' clustering following the abundance of each metabolic pathway predicted for each sample.
To assess functional differentiation among the different treatments (per type of sample), LEfSe was performed, using the online tool Microbiome Analyst (https://www.microbiomeanalyst.ca/; Chong et al., 2020; Dhariwal et al., 2017) using the same parameters as for the bacterial community analysis (see above).
3 RESULTS
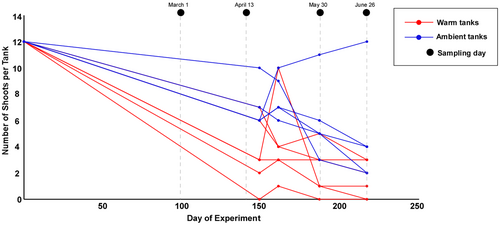
The overall trend observed in the number of Z. marina shoots registered throughout the experiment indicated a decrease from the beginning of the sampling period to the end of the study (Figure 2).
After quality control, removal of chloroplast and mitochondria, as well as singletons and doubletons a total of 1,037,644 sequences corresponding to 3140 ASVs (https://doi.org/10.5281/zenodo.11132203) were obtained. To account for variation in sequence depth among samples (ranging from 3619 to 22,256 sequences), rarefaction was performed to normalize to the minimum number of sequences (3619). The rarefied dataset contained 3126 unique ASVs and was used for all further statistical and multivariate analyses.
3.1 Bacterial community diversity and structure
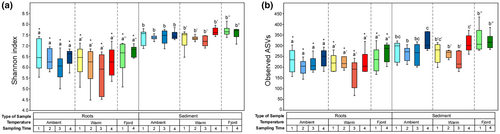
To assess bacterial community diversity and evenness (alpha diversity), we calculated the Shannon diversity index and the number of observed ASVs for each sample. Overall, sediments had a consistently higher diversity than root communities regardless of the factor considered (Figure 3a,b). Differences between the two sample types ranged from a to b. Within root communities, both diversity indices showed no differences among the different sampling times (1, 2, 3, and 4) within each temperature setting/treatment (Ambient, Warming, and Fjord [Control]; Figure 3a). Similarly, no differences were found among the different temperature treatments for the same sampling time (Figure 3a). For the sediments, the Shannon index did not reveal any difference among samples (Figure 3a). However, the number of ASVs within the Ambient setting was higher for samples collected during sampling time 4 compared to sampling time 2 (Figure 3b). In the Warm treatment, samples from the sampling times 2 and 3 exhibited a lower diversity as compared to sampling time 4 (Figure 3b). Within the Fjord, no diversity differences were found among the sampling times.
The increase in seawater temperature affected the structure of microbial communities depending on both sample type (sediment or root) as well as sampling time. Among the single factors, the sample type (p = .001, Table 1, Table S1, Figure 4) accounted for most variation among samples (Table 1). Temperature treatment and sampling time as single factors contributed equally to community structure by around 10% (Table 1).
| Source of variation | df | SS | MS | Pseudo-F | p (perm)a | Variation explained (%) |
|---|---|---|---|---|---|---|
| Type of sample (Ty) | 1 | 69,725 | 69,725 | 33.965 | .001 | 22.35 |
| Temperature treatment (Te) | 2 | 24,366 | 12,183 | 5.9347 | .001 | 9.95 |
| Sampling time (Sa) | 3 | 26,674 | 8891.4 | 4.3313 | .001 | 9.28 |
| Ty × Te | 2 | 13,898 | 6949.1 | 3.3852 | .001 | 9.78 |
| Ty × Sa | 3 | 12,707 | 4235.6 | 2.0633 | .001 | 7.42 |
| Te × Sa | 4 | 18,271 | 4567.7 | 2.2251 | .001 | 8.59 |
| Ty × Te × Sa | 4 | 10,660 | 2664.9 | 1.2982 | .001 | 5.99 |
- Note: H0: there are no differences in the distribution of ASVs for the different treatments, H0 rejected if p < .05. p-values based on 999 permutations.
- a PERMANOVA values were calculated after square root transformation and Bray–Curtis distances calculation for ASV composition.
When comparing the different temperature regimes (Ambient, Warm, and Fjord control) for each sampling time (roots and sediments separately), bacterial communities were always statistically different, except when comparing those associated with roots sampled in sampling time 1 and sediments sampled in the first and second dates (Table S2a,b).
As for the bacterial community changes across the different sampling times, for each temperature treatment, the microbiome associated with roots and sediments at ambient temperature did not significantly change from sampling time 1 to sampling time 2 (Table S3a). However, at warm treatment, only the microbiome associated with the roots sampled in sampling time 4 was different from the remaining (Table S3b) but for the sediments, as before, bacterial communities were always statistically different except between sampling 1 and 2 (Table S3b). The samples collected in the Fjord have also shown differences in the bacterial community associated with roots and sediments sampled in the two different sampling times (Table S3c).
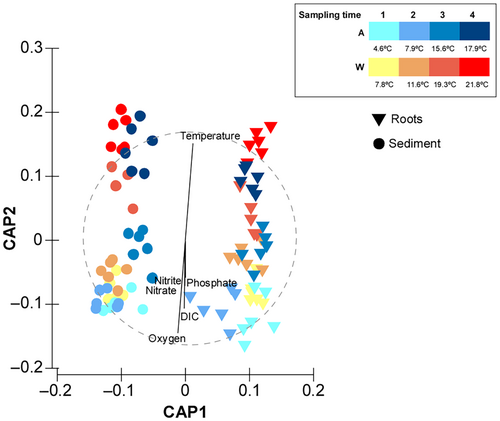
The PERMANOVA outcomes that are shown in the CAP display a clear and strong separation between roots and sediments (Figure 4) but, in addition we can see a separation by sampling point reflecting a separation driven by actual temperature (Figure 4), which is especially clear in the sediments.
The abiotic factors measured in this study were overlayed in our multivariate analysis, showing that temperature and oxygen had the strongest correlation to the multivariate patterns separating bacterial communities at different temperatures (Figure 4).
3.2 Microbial community composition and main shifts across temperature treatments and temperature gradients
To ensure a reliable analysis of bacterial community composition for each treatment, we removed the ASVs that were not present in at least 80% of the replicates.
3.2.1 Bacterial community associated with Z. marina roots
Overall, the bacterial communities associated with Z. marina roots were distinct from those at the collection site (Fjord). However, samples from the Ambient treatment at sampling time 1 and 4 share a higher number of taxa with the correspondent Fjord samples (for sampling time 1 no discriminant features were found between these two treatments), including two of the most abundant taxa (Spirochaetaceae and Gammaproteobacteria for sampling time 1 and Gammaproteobacteria and Sulfurimonadaceae for sampling time 4; Figure 5a,c). For the fjord roots, sampled at the end of the experiment, Arcobacteraceae and Bacteroidetes BD2-2 were found as discriminant when comparing to ambient samples from that same period (Figure S1a).
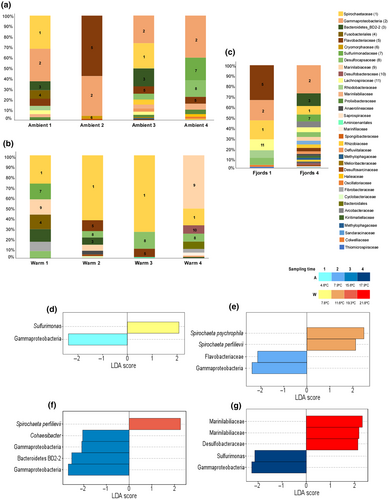
In the Ambient treatment, there was a decrease in the number of characterizing taxa at the sampling time 2, Flavobacteriales and Gamaproteobacteria dominated the total taxa relative abundance (Figure 5a). However, by sampling time 3 the bacterial community reverted to a bacterial community more similar to sampling time 1, sharing three of the most abundant taxa (Spirochaetaceae, Gammaproteobacteria, and Bacteroidetes BD2-2; Figure 5a). At sampling time 4, the Gammaproteobacteria was still the most abundant taxon, but there was also an increase of members from the Sulfurimonadaceae and Desulfocapsaceae families, which were within the three most abundant for this treatment (Figure 5a).
In the Warm treatment, there was an increase in the Spirochaetaceae (the most abundant taxa for the three first sampling times), along with temperature (Figure 5b). At the highest temperature (sampling point 4) the most abundant taxa were identified as Marinilabiliaceae. In contrast to the ambient treatment, at least one taxon related to the sulfur cycle was among the most abundant across the experiment in the Warm treatment. In the warmest sampling time, two, out of four, different families of sulfate-reducing bacteria (Desulfocapsaceae and Desulfobacteraceae; Figure 5b) were among the most abundant.
When comparing corresponding sampling times between the two temperature treatments (Ambient and Warm), we found that the higher the temperature the greater the number of biomarkers/discriminant bacteria between the homologous sampling times (Figure 5d–g). At sampling times 1, Gammaproteobacteria was the discriminant taxa for the Ambient treatment while Sulfurimonas was the discriminant taxa for the Warm treatment (Figure 5d). Two different Spirochaeta species were discriminant for the second sampling in the Warm treatment, while Flavobacteria and Gammaproteobacteria are more abundant in the Ambient treatment (Figure 5e). Two ASVs identified as Gammaproteobacteria were found to be discriminant for the third sampling at Ambient treatment, along with Bacteroidetes BD2-2 and Cohaesibacter, but the Warm treatment was still represented by a Sprirochaeta (Figure 5f). In the last sampling time, Sulfurimonas and Gammaproteobacteria were significantly more abundant at the Ambient temperature while the Warm treatment was discriminated by two ASVs identified as Marinilabiliaceae and a Desulfobacteraceae (Figure 5g).
3.2.2 Bacterial community associated with bulk sediments
The sediment-associated microbiome was more stable compared with the root-associated community, and this was observed across all three treatments (Fjord/Control, Ambient, and Warm; Figure 6a–c). Although sharing the most abundant taxa, the ambient sediments collected in the first sampling day showed a lower number of taxa when compared to the first fjord sediments (Figure 6a,c). Also, winter fjord samples showed a higher number of discriminant taxa (e.g., Actinomarinales, Bacteroidales SB-5, Thiogranum, Lutimonas; Figure S1b). Comparing the latest sampling time, the major taxonomic groups changed and only Flavobacteriaceae was shared among the most abundant taxa for ambient and fjord sediments (Figure 6a,c). Actinomarinales, Chromatiaceae, Bacteroidetes VC2.1 Bac.22, Thiomicrospiraceae, and Eudoraea were within the discriminant taxa for fjord sediments and Microscillaceae, Oscilatoriaceae, and Candidatus Thiobius differentiate the ambient samples (Figure S1c).
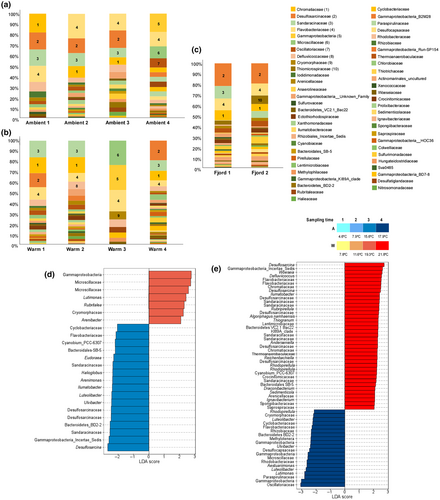
The microbiomes of the Ambient temperature treatment shared the same four most abundant taxa (Chromatiaceae, Desulfosarcinaceae, Sandarinaceae, and Flavobacteriaceae; Figure 6a), until the fourth sampling when a major shift in the microbiome occurs, and the most abundant taxa are members of the Gammaproteobacteria, Flavobacteriaceae, Microscillaceae, and Oscillatoriaceae (Figure 6a).
The Warm treatment presented the same most abundant taxa as the ambient treatment in the first sampling time and in the second sampling only the Desulfosarcinaceae were replaced by the Defluviicoccaceae (Figure 6b). In the third sampling point, there was a strong shift in the bacterial community associated with the sediments, where the most abundant taxa were Flavobacteriaceae, Gammaproteobacteria, Microscillaceae, and Cryomorphaceae (Figure 6b). In the last sampling, the bacterial community apparently reverted to the initial most abundant taxa (Chromatiaceae, Desulfosarcinaceae, Sandarinaceae, and Flavobacteriaceae).
Ambient and Warm treatments had significant bacterial biomarkers/discriminants for sampling times 3 and 4 (Figure 6d,e). For sampling time 3 in the Ambient treatment, 17 taxa biomarkers were identified, including several genera belonging to less abundant families that were mostly found among Ambient temperature replicates (e.g., Halioglobus, Arenimonas, Ilumatobacter, and Luteolibacter) (Figure 6d). Within the discriminant taxa were also bacteria from the Sandaracinaceae, Desulfosarcinaceae, and Flavobacteriaceae families (Figure 6d). For the Warm treatment, 7 biomarkers taxa were identified, and those with higher LDA scores belonged to the Gammaproteobacteria and Microscillaceae family (Figure 6d). For the sampling time 4, 20 Ambient biomarkers were found and the ones with the highest LDA score belonged to Gammaproteobacteria and the Oscillatoriaceae, Rhodobacteraceae, Microscillaceae families (Figure 6e). In sediments sampled at the highest temperature, 39 taxa were detected as discriminants, including a great proportion of taxa from the Desulfosarcinaceae, Chromatiaceae, Sandarinaceae, and Flavobacteriaceae families (Figure 6e) as well as several ASVs within less abundant families not found in the Ambient treatment (e.g., Thermoanaerobaculaceae).
3.3 Analysis of the predicted functional profiles—PiCRUSt
We conducted the PERMANOVA analyses separately for each type of sample (sediment and root) due to the strong signal between roots and sediments (p < .001) that would mask the differences among the other factors, the Fjord samples were not included in these statistics as we were mainly interested in the differences among the different treatments.
For both roots and sediments, a significantly different putative functional profile was found among the different sampling times, irrespective of temperature treatment (p = .031//.008, Table 2, Figure 7). The functional profiles were mostly clustered based on the effective temperature at which samples were collected. For the roots, a major cluster with samples from sampling times 3 and 4 (A and W treatments) while the earliest sampling times are more dispersed. However, we only found statistical differences between sampling 3 and the remaining (see Table S4) which would be mostly due to the ambient samples that cluster apart in the CAP plot (Figure 7a). For the sediments, the clustering is more evident for sampling times 1 and 2 (A and W treatments) which did not differ among each other (Figure 7b, p = .407, Table S4). The other cluster represents samples collected at temperatures higher than 15.6°C where only sampling time 3 statistically differs from sampling 2 and 4 (Figure 7b, Table S4).
| Source of variation | SS (roots//sediments) | MS (roots//sediments) | Pseudo-F (roots//sediments) | p (perm)a (roots//sediments) |
|---|---|---|---|---|
| Temperature treatment (Te) | 386.89//36.47 | 386.89//36.475 | 1.4629//0.3741 | .214//.607 |
| Sampling time (Sa) | 2326.70//1157.10 | 775.56//385.71 | 2.9326//3.9555 | .031//.008 |
| Te × Sa | 515.15//444.48 | 171.72//148.16 | 0.6493//1.5194 | .621//.236 |
- Note: H0: There are no differences in the distribution of OTUs for the different treatments, H0 rejected if p < .05. Boldface values are significant at p < .05, p-values based on 999 permutations.
- a PERMANOVA values were calculated after square root transformation and Bray–Curtis distances calculation for ASV composition.
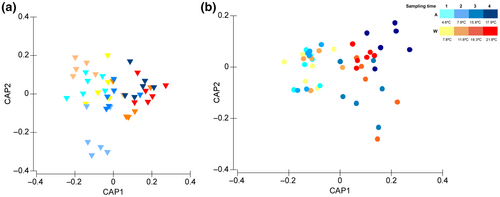
To identify potential key functions that would allow Z. marina to thrive under higher temperatures, we have only presented those functions that were found significantly more abundant at higher temperatures (considering the clustering above—Figure 7—we have considered sampling times 3 and 4, from both temperature setting, as the warmest treatments) (Figure 8a,b).
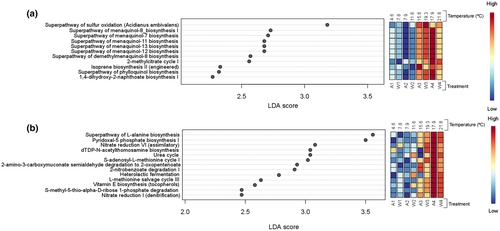
For root-associated microbial communities, 11 biomarkers were consistently and significantly more abundant in warmer treatments (temperatures ≥15.6°; Figure 8a). These were: Superpathway of sulfur oxidation, superpathway of menaquinol (−7, 8. 11, 12, 13) biosynthesis, superpathway of demethylmenaquinol-8 biosynthesis, 2-methylcitrate cycle I, Isoprene biosynthesis II, superpathway of phylloquinol biosynthesis and 1,4-dihydroxy-2-naphtoate biosynthesis I.
For the microbial communities of sediments, 13 biomarkers highly abundant among higher temperatures were found, which were: Superpathway of l-alanine biosynthesis, pyridoxal-5 phosphate biosynthesis I, nitrate reduction VI (assimilatory) and I (denitrification), dTDP-N-acetylthomosamine biosynthesis, urea cycle, S-adenosyl-l-methionine cycle I, 2-amino-3-carboxymuconate semialdehyde degradation to 2-oxopentenoate, 2-nitrobenzoate degradation I, heterolactic fermentation, l-methionine salvage cycle III, vitamin E biosynthesis (tocopherois), and S-methyl-5-thio-alpha-d-ribose 1-phosphate degradation (Figure 8b).
4 DISCUSSION
This study is the first to experimentally investigate the impact of a chronical increase in SST, through a 9-month trans-seasonal period, on the bacteria associated with the coastal temperate seagrass Z. marina roots and bulk sediments in the Baltic Sea. Our aim was to evaluate how persistent elevated temperature would affect the microbial community associated with this foundation seagrass across more than three seasons. In general, the effects of temperature emerged earlier in the warming treatment with an increase in organic matter-degrading and sulfur-related bacteria. In the warmest months of the warm treatment, Z. marina roots' bacterial community has shown a higher ratio of sulfate-reducing bacteria compared to sulfide oxidizers. Also, the differentiating predicted pathways were mostly related to biogeochemical cycles and possible production of hydrocarbons that would protect seagrass from temperature stress.
4.1 Bacterial communities associated with Z. marina roots
4.1.1 Shifts through seasons under ambient temperature
The bacterial community of ambient roots sampled in winter (sampling point 1) differed from that of the control samples (Fjord), which could be related to the transplant and acclimation process, where the individuals did not entirely switch back to their original microbiome. However, none of the taxa were significantly discriminant when comparing both samples (LEfSe analysis). Spirochaetaceae and Gammaproteobacteria, two of the most abundant taxa, were common to both treatment groups. Gammaproteobacteria are a large group of metabolically different bacteria and Spirochaetaceae have been linked to cell wall degradation in seagrass ecosystems (Trevathan-Tackett et al., 2017), involved in the fixation of nitrogen and carbon (Van De Water et al., 2016), and commonly associated with seagrass meadows (Hurtado-McCormick et al., 2019). For the summer sampling, the fjord roots showed a significantly higher abundance of sulfur oxidizers (SOs; Arcobacteriaceae) and Bacteroidetes BD2-2 which are known to contribute to the carbon cycling and have also been associated with healthy seagrass meadows with a dense shoots' coverage (Randell et al., 2023). The higher accumulation of sulfide in the ambient treatment, compared with the fjord/control, might be inhibiting the carbon cycling and consequently the bacteria associated with that cycle (Wang, Tomas, et al., 2020; Wang, Woo, et al., 2020).
Contrarily to what happened when considering the total bacterial community, the core microbiome of ambient roots sampled during the winter differed from those sampled in early spring. Flavobacteriales and Gamaproteobacteria took over most of the total taxa relative abundance in the roots sampled in early spring (sampling time 2). Flavobacteriaceae are mostly known for their efficiency at initial organic matter degradation (Bissett et al., 2008). This increase could have already been an early indication of the health state of the shoots that might have been susceptible to some degradation. A decrease in the number of shoots was registered in the next months.
Reinforcing the idea that the microbiome shift from winter to early spring ambient roots was not triggered by the increasing temperature, the bacterial community associated with late spring ambient roots reverted back to a bacterial community more similar to that of the first sampling time, sharing three of the most abundant taxa (Spirochaetaceae, Gammaproteobacteria, and Bacteroidetes BD2-2). The microbiome of samples from the ambient roots sampled during summer, showed a new increase in bacteria from the Gammaproteobacteria phylum. Also, two sulfur cycle-related taxa were among the most abundant (Sulfurimonadaceae and Desulfocapsaceae). After an abnormally warm June, Sawall et al. (2021) registered a comparatively high mortality of plants at the end of the summer. Although our experiment took place in adjacent aquaria within the same tank as Sawall et al.'s (2021), and while we did not directly record mortality, we observed a reduction in the number of shoots in our aquaria. This might explain the increased abundance of Gammaproteobacteria degraders (compared with the previous sampling) with a consequent activation of the sulfur cycle. In this treatment, a balanced abundance of sulfate-reducing bacteria (Desulfocapsaceae) and sulfur oxidizers (Sulfurimonadaceae) would help in sulfide detoxification (Koch, Schopmeyer, Kyhn-Hansen, et al., 2007) as observed before in “stressed” Halophila ovalis (Martin et al., 2020).
4.1.2 Shifts through seasons under chronical warming
In Sawall et al. (2021) experiment, the warm treatment resulted in a strong selection for heat-resistant shoots, with a peak of mortality occurring in early spring (in between the second and third sampling time). We have also registered a decrease in the number of shoots in that same time period. This likely led to an increase in organic material which, in turn, contributed to the heightened abundance of Spirochaetacea in the first three sampling points. Spirochaetacea are key degraders and decomposers of complex organic material commonly found in the seagrass rhizosphere (Wang et al., 2021). In the summer, Marinilabiliaceae were found to be the main degraders in Z. marina roots, as they can metabolize different forms of organic carbon (Conte et al., 2023; Tu et al., 2022). Unlike the ambient setup, that early temperature effect has also stimulated the sulfur cycle throughout the setup, with a stronger increase of sulfate reducers as temperature increases. In the summer, we could find two different families of sulfate-reducing (SR) bacteria, Desulfocapsaceae and Desulfobacteraceae—Thermodesulfobacteriota that thrive at higher temperatures (Lobo et al., 2012)—in opposition to the homologous sampling time in the Ambient treatment where we found a similar proportion of sulfate reducers and SO. Different studies on several seagrass species (Brodersen et al., 2024; Martin et al., 2019; Rolando et al., 2022), sulfide oxidizers not only aid in eliminating toxic sulfide from the root area but also facilitate N and/or C fixation through their oxidation of sulfide. An unbalanced ratio of SR:SO can lead to toxic levels of sulfide which could eventually collapse the seagrasses' population if not effectively removed. In a nutrient enrichment mesocosms experiment, by Wang, Tomas, et al. (2020) and Wang, Woo, et al. (2020), excess of nutrients led to an increase in sulfur and nitrogen bacteria in Z. marina belowground communities. This suggests that nutrient enrichment can stimulate biogeochemical cycling, potentially exacerbating sulfide toxicity in sediments.
In the warm treatment, a possible accumulation of detritus/organic material, coupled with high temperatures, may have resulted in a higher mortality rate at the end of the experiment, as reported by Sawall et al. (2021) and corroborated by a huge decrease in the number of shoots in our aquaria. This trend has also been observed in other studies that showed a decline in cold-affinity seagrass populations, like Z. marina, associated with increased sulfate reduction rates and consequent sulfide accumulation resulting from increased SST (García et al., 2012, 2013; Kim et al., 2017; Koch, Schopmeyer, Holmer, et al., 2007). In adult plants, the main signs of heat stress, such as the loss of biomass, are typically visible only after several weeks (Reynolds et al., 2016).
4.1.3 Main shifts between ambient and warm setups
Through differential analysis of homologous sampling times from different temperature treatments, certain taxa were found to be differentially correlated with higher temperatures. The differentially abundant taxa were primarily organic matter degraders, such as Gammaproteobacteria and Flavobacteriaceae for the ambient treatment and Spirochaetaceae and Marinilabiliaceae for the warm benthocosms. A possible increase of organic matter resulting from Z. marina mortality, a side effect of the increased SST, is likely the main driver of this bacterial shift. This differentiation of degrading taxa may be related to seawater temperature and the different tolerances of the differentiated taxa. For example, Spirochaetaceae have been found to be associated with corals that were more resistant to heat stress than those that were more susceptible (Ziegler et al., 2017).
In addition, differential taxa related to the sulfur cycle were identified, with Sulfurimonas being differentially more abundant in warm roots (WR) sampled in winter and Desulfobacteraceae being the differentially most abundant taxa in summer WR. As expected, in the warm treatment there is a stimulation of sulfur cycling. Most seagrasses can tolerate low concentrations of the toxic sulfide by H2S oxidation (Hasler-Sheetal & Holmer, 2015). Bacteria as the above-mentioned Sulfurimonas are one of the taxa responsible for that detoxification and its presence in the core microbiome might be vital for seagrass survival under environmental stressors (de la Garza Varela et al., 2023; Wang et al., 2021). However, when the sulfate reduction surpasses sulfide oxidation the level of sulfides can get very high and its intrusion into seagrass tissues can lead to massive die-off events (García et al., 2012, 2013; Holmer et al., 2009). So, finding Desulfobacteraceae as the main biomarker for the warmest samples can be an indication that sulfides may have reached detrimental levels. As discussed before, two different sulfate reducer taxa (Desulfocapsaceae and Desulfobacteraceae) were found as the most abundant in summer WR which highlights a possible stronger sulfate reduction in samples from this treatment compared to the other treatments. These results are consistent to the ones from Kardish and Stachowicz (2023). They suggest that members of Desulfocapsaceae (with conserved sulfur metabolism), found associated with Z. marina roots in a warmer environment, are adapted to warmer temperatures and could be good indicators of environmental changes.
4.2 Bacterial communities associated with bulk sediments
4.2.1 Shifts through seasons under ambient temperature
The fjord sediments sampled in both winter and summer, when compared to the homologous ambient sediments, showed a higher abundance of Actinomarinales and Bacteroidetes SB-5 and VC2.1 Bac22. Actinomarinales, known as key heterotrophs for carbon mineralization (Miksch et al., 2021), have been found associated with sediments characterized by greater vegetation coverage (Randell et al., 2023). That is in accordance with the low abundance of shoots at the end of the experiment. Bacteroidetes VC2.1 Bac22 have been associated with complex organic carbon mineralization (Leng et al., 2022). Once more, like for the roots, the carbon cycle might have been affected in the ambient setup due to the higher accumulation of sulfide (Wang, Tomas, et al., 2020; Wang, Woo, et al., 2020). Also, plants' resistance to environmental stresses have been associated with a reduction in sediment-associated Bacteroidetes (Pérez-Jaramillo et al., 2018). So, again, the control samples, in both sampling times, show a higher abundance of taxa that are usually associated with healthier environments.
Compared with the roots, the core community of each sediment treatment showed a higher number of associated taxa. However, the four most abundant taxa were kept throughout most of the experiment, suggesting that under stress, sediments are more stable than the plant, as previously proposed (Markovski et al., 2022). The most abundant taxa found in Z. marina bulk sediments (Chromatiaceae, Desulfosarcinaceae, Sandaracinaceae, and Flavobacteriaceae) are commonly found in the seagrass core rhizosphere (Cúcio et al., 2016, 2018; de la Garza Varela et al., 2023; Ettinger et al., 2017; Wang et al., 2021). Members of the Chromatiaceae family are mainly SOs and can actively consume the sulfide from the rhizosphere, contributing to the detoxification of the root area (Cúcio et al., 2018). Desulfosarcinaceae are sulfate-reducing bacteria (SRB) that produce the phytotoxic hydrogen sulfide which will be released in the sediments.
Despite the apparent stability across the seasons, in the ambient warmest season, the most abundant taxa shifted to a different composition. There was a high growth of cyanobacteria from the Oscillatoriaceae family which may have profited from the increasing nutrients resulting from organic matter degradation (Brocke et al., 2015). This increase might explain the high abundance of Microscillaceae which have been described as a predator (Chun et al., 2020) and an obligate parasite for cyanobacteria (Daft & Stewart, 1971).
4.2.2 Shifts through seasons under chronical warming
The first sampling period of the warmed sediments (WS) revealed the same four most abundant taxa observed in the ambient setup (Chromatiaceae, Desulfosarcinaceae, Sandaracinaceae, and Flavobacteriaceae). However, a possible mortality peak in early spring as registered by Sawall et al. (2021), which in our sampling set resulted in a decrease of the number of shoots, may have led to that earlier community changes detected in the second and third sampling periods. In the early spring WS samples, one of the most abundant taxa was assigned to the Defluviicoccaceae family, reported to catalyze denitrification (Wang, Tomas, et al., 2020; Wang, Woo, et al., 2020). This process may have been stimulated by the accumulation and degradation of organic matter, resulting in a high concentration of nutrients. Cryomorphaceae were also identified as one of the most abundant taxa in these samples. Members of this family are largely dependent on decaying organic biomass (Bowman, 2020). The sediment samples, collected in the summer and warmest treatment, presented a microbiome with the same four most abundant taxa observed in the winter samples. This reinforces the idea that most of the changes in the sediment microbiome were likely triggered by organic matter accumulation rather than by high temperatures, directly.
4.2.3 Main shifts between ambient and warm setups
In contrast to the roots, differences between ambient and warm treatments in the sediments were only observed in the warmest sampling times (3 and 4), indicating a higher stability. In the third sampling time, the ambient treatment had more sulfur reducers as differential biomarkers compared to the warming setup, which showed the presence of the cyanobacteria pathogen Microscillaceae and the decaying organic biomass-dependent Cryomorphaceae. In general, a high abundance of SRB, SO, and organic matter degraders are common in the sediment microbiome under “normal conditions” (Cúcio et al., 2018). However, in WSs sampled in late spring, the abundance of these taxa was disrupted most likely due to the increased mortality of Z. marina shoots and its decay, leading to the emergence of differential taxa related to that circumstance. Similarly, when comparing ambient and warmed summer sediments, the warmest treatment which have already reverted back to its most abundant taxa to the initial composition, shows SRB and SO as the most common biomarkers while taxa related to biomass degradation are the differential taxa in summer ambient sediments, where Z. marina high mortality occurred.
4.3 Predicted functional profiles differentiation between ambient and warm treatments
The overall analysis of predicted functions for roots and sediments microbiomes showed, in contrast to the microbial community structure, minimal differentiation among treatments. This may be due to different bacterial taxa performing similar functions throughout the experiment (Chen et al., 2022). However, specific differences were observed in the functional profiles of the warmest months (sampling points 3 and 4 for both ambient and warm treatments). The superpathway of sulfur oxidation and isoprene biosynthesis were identified as the primary biomarkers for the roots sampled during the warmest months. The former suggests increased sulfur cycle activity in warmer treatments, consistent with the high abundance of sulfur cycling-related bacteria in the core community structure, most of them thermophilic or moderate thermophilic (Nakagawa & Takai, 2008). Furthermore, as previously discussed, elevated SST can lead to the die-off of sensitive marine organisms, including seaweeds and seagrasses, resulting in increased organic matter availability and stimulation of sulfur-related bacteria (Duarte et al., 2018; Rolando et al., 2022).
The latter, isoprene biosynthesis II, is, in some terrestrial plants, linked to the production of this volatile organic compound that protect against thermal shock and oxidative stress (Medori et al., 2012). These compounds can be produced by bacteria to enhance resistance in their host plants (McGenity et al., 2018). In addition, several pathways related to menaquinone biosynthesis were increased in warmer treatments. Menaquinone is a vital component of electron transport chains in many microorganisms and may be involved in energy production (Dairi, 2012). This suggests that the microbiome of Z. marina roots increased its activity in response to an environmental stressor such as warming temperatures, enabling a rapid response (Sarmento et al., 2010).
The pathways that better differentiated the warmer treatments in sediments were nitrate reduction (both assimilatory and denitrification) and heterolactic fermentation. Maximum activity of nitrogen removal processes occurs during warmer months in the Baltic Sea, when sediment organic content is higher (Jäntti et al., 2011). Another study reported a 50% increase in nitrate uptake by Z. marina under extreme warming scenarios (Alexandre et al., 2020; Kaldy, 2014), indicating that this mechanism plays a critical role in eliminating excess nutrients from water and helping plants adapt to chronic warming (Dai et al., 2020; Qu et al., 2022). As previously discussed, Z. marina die-off increased organic matter in the sediments, leading to increased decomposition and fermentative processes.
4.4 Conclusions
This study suggests that the effects of temperature emerged earlier in the superimposed warming treatment compared with the natural seasonal warming. The main differences are most likely due to premature Z. marina die-offs under chronic warming conditions, resulting in an earlier increase in organic matter-degrading bacteria and a higher ratio of sulfate-reducing bacteria compared with sulfide oxidizers. Also, the differentiating predicted pathways were mostly related to biogeochemical cycles and possible production of hydrocarbons that would protect seagrass from temperature stress.
These structural and compositional variations of the associated microbiome can provide insights into the seagrass ecological status, and some taxa/genes/pathways may be used as markers for specific stresses. Conte et al. (2021) proposed that taxa engaged in crucial biogeochemical transformations, particularly those related to nitrogen, sulfur, and carbon cycles, would be the most effective indicators. Any changes in these markers can impact the ecology and physiology of the seagrass host. To further understand these fluctuations in the seagrass holobiont under the influence of climatic stressors, controlled laboratory research, such as in the present study, is essential. Seagrass monitoring and conservation programs should integrate results from such research to identify early warning indicators of the seagrass physiological state and prevent the extinction of vulnerable populations. Additionally, a better understanding of how the microbiome can change under the influence of certain stresses is essential to investigate specific taxa that might function as probiotics and be used to prevent the major effects of climate change or restore seagrass ecosystems.
AUTHOR CONTRIBUTIONS
Tânia Aires: Data curation; formal analysis; investigation; methodology; validation; visualization; writing – original draft; writing – review and editing. Catarina Cúcio: Data curation; formal analysis; methodology. Janina Brakel: Data curation; investigation; methodology; writing – review and editing. Florian Weinberger: Conceptualization; data curation; formal analysis; funding acquisition; investigation; methodology; project administration; validation; writing – review and editing. Martin Wahl: Conceptualization; data curation; funding acquisition; investigation; project administration; resources; validation; writing – review and editing. Ana Teles: Methodology; writing – review and editing. Gerard Muyzer: Formal analysis; investigation; methodology; writing – original draft; writing – review and editing. Aschwin H. Engelen: Conceptualization; formal analysis; methodology; supervision; writing – original draft; writing – review and editing.
ACKNOWLEDGEMENTS
We are very grateful to all the people who helped in the sampling process by providing all the means and logistics to make it possible. This project was partly funded by GEOMAR and by the Cluster of Excellence “The Future Ocean.” The “Future Ocean” is funded within the framework of the Excellence Initiative by the Deutsche Forschungsgemeinschaft (DFG) on behalf of the German federal and state governments. This study also received Portuguese national funds from FCT—Foundation for Science and Technology through projects UIDB/04326/2020, UIDP/04326/2020 and LA/P/0101/2020. A fellowship to TA (SFRH/BPD/116774/2016) and contract CEECINST/00114/2018 to AE. CC and GM were funded by a grant of the European Union (MaCuMBA). In addition, GM was funded by the research priority area System Biology of the University of Amsterdam. JB was funded by a PhD scholarship through “Deutsche Bundesstiftung Umwelt” (DBU).
CONFLICT OF INTEREST STATEMENT
The authors declare no conflicts of interest.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are openly available in Zenodo database at https://doi.org/10.5281/zenodo.11132203.




