Arctic plasmidome analysis reveals distinct relationships among associated antimicrobial resistance genes and virulence genes along anthropogenic gradients
Abstract
Polar regions are relatively isolated from human activity and thus could offer insight into anthropogenic and ecological drivers of the spread of antibiotic resistance. Plasmids are of particular interest in this context given the central role that they are thought to play in the dissemination of antibiotic resistance genes (ARGs). However, plasmidomes are challenging to profile in environmental samples. The objective of this study was to compare various aspects of the plasmidome associated with glacial ice and adjacent aquatic environments across the high Arctic archipelago of Svalbard, representing a gradient of anthropogenic inputs and specific treated and untreated wastewater outflows to the sea. We accessed plasmidomes by applying enrichment cultures, plasmid isolation and shotgun Illumina sequencing of environmental samples. We examined the abundance and diversity of ARGs and other stress-response genes that might be co/cross-selected or co-transported in these environments, including biocide resistance genes (BRGs), metal resistance genes (MRGs), virulence genes (VGs) and integrons. We found striking differences between glacial ice and aquatic environments in terms of the ARGs carried by plasmids. We found a strong correlation between MRGs and ARGs in plasmids in the wastewaters and fjords. Alternatively, in glacial ice, VGs and BRGs genes were dominant, suggesting that glacial ice may be a repository of pathogenic strains. Moreover, ARGs were not found within the cassettes of integrons carried by the plasmids, which is suggestive of unique adaptive features of the microbial communities to their extreme environment. This study provides insight into the role of plasmids in facilitating bacterial adaptation to Arctic ecosystems as well as in shaping corresponding resistomes. Increasing human activity, warming of Arctic regions and associated increases in the meltwater run-off from glaciers could contribute to the release and spread of plasmid-related genes from Svalbard to the broader pool of ARGs in the Arctic Ocean.
1 INTRODUCTION
The emergence and spread of antimicrobial resistance (AMR) is among the greatest threats to global health (World Health Organization [WHO], 2021). Widespread use and misuse of antimicrobials in humans, farm animals and crop production increases the selection pressure for the survival of resistant strains and can contribute to the mobilization of antibiotic resistance genes (ARGs) via horizontal gene transfer (HGT). Other antimicrobials, such as heavy metals, are also commonly found in the environment and can impose similar ecological pressures (Levy & Marshall, 2004; Wales & Davies, 2015). Whilst most efforts to stem the spread of AMR have focused on the human health and agriculture sectors, it is now clear that important environmental dimensions are also at play. Both biological (e.g., application of human/animal fecal material) and chemical (heavy metal contamination) inputs to affected environments, including water and soil, can contribute to the transmission and spread of AMR (Perry & Wright, 2013). Additionally, climate change—in particular increasing temperatures—could increase microbial growth rates and expand niches available for pathogen survival, whilst increased frequency and intensity of storms, and flooding, as well as increased global human traffic, could contribute to the increased dispersal of various resistant bacteria and their ARGs to foreign environments (Frost et al., 2019; Global Leaders Group [GLG] on Antimicrobial Resistance [AMR], 2021).
The diversity and abundance of ARGs in both anthropogenically influenced as well as pristine environments suggests that reservoirs of ARGs are diverse and widespread (Berendonk et al., 2015; Li et al., 2022; Makowska et al., 2020; Makowska-Zawierucha et al., 2022). Pristine environments are especially pertinent for assessing the natural (i.e., not anthropogenic) state of resistance before the modern mass production and release of antibiotics, antimicrobials and other pollutants (Allen et al., 2009; Pruden et al., 2006). However, even remote environments, such as Arctic glaciers and fjords, are now also being found to be polluted by heavy metals and organic compounds (Łokas et al., 2016; Makowska-Zawierucha et al., 2022). Recent studies have, for instance, indicated the occurrence of various pollutants originating from both local (such as industry, tourism and scientific activity) and distant sources (Gorecki et al., 2021; McCann et al., 2019). The Arctic environment therefore provides the opportunity to observe the combined effects of increasing human activity and climate change on AMR spread. The intersections of naturally occurring phenomenon (including distinct seasonality and the escalation of biological activity during summer) and anthropogenic factors (such as climate change and the intensifying tourism industry in the Arctic) make the study of AMR spread in the Arctic particularly vital.
Global warming is inducing widespread glacial melting and retreat, which has the potential to release ancient (from old ice layers) or modern (from the snow overlaying glaciers) genetic diversity to the environment (Mao et al., 2023; Segawa et al., 2013). At the same time, wastewater discharge and other human inputs also release microbes and other pollutants such as biocides and metals to Arctic fjords and other downstream environments, in turn increasing the gene pool of the marine resistome (Larsson & Flach, 2022; Makowska-Zawierucha et al., 2022; Pouch et al., 2023; Sajjad et al., 2020). Meanwhile, the so-called Atlantification of the Arctic (the ingress of warm ocean water currents into Arctic waters including oceans and fjords) will likely transport biotic and abiotic elements including pollutants from low latitude and boreal regions to pristine high-latitude ecosystems (Csapó et al., 2021; Wichmann et al., 2019). Ocean currents could correspondingly facilitate the global transport of antibiotic resistance (Yang et al., 2021), as has been reported with microplastic particles—which provide a vehicle for dispersal of drug-resistant bacteria carried by ocean currents to the deep sea (Stenger et al., 2021).
Horizontal gene transfer encompasses three main processes by which bacteria can share their genes and acquire novel traits, specifically: transformation (the uptake of free DNA), transduction (gene transfer mediated by bacteriophages) and conjugation (gene transfer by means of plasmids or integrative conjugative elements) (Frost et al., 2005; Rodríguez-Beltrán et al., 2021). Mobile genetic elements (MGEs), DNA elements that can move within genomes or between bacterial cells; such as plasmids, bacteriophages, integrative conjugative elements, transposons, insertion sequence (IS) elements, gene cassettes and genomic islands, are key drivers of HGT (Figure 1) (Frost et al., 2005; Rodríguez-Beltrán et al., 2021). Plasmids are of particular interest because of the role that they play in disseminating ARGs between and across species, including pathogens (Levy & Marshall, 2004; Martínez et al., 2015; Stalder et al., 2019). Plasmids are autonomously replicating circular or linear DNA molecules that can stably coexist with chromosomes and that commonly carry ARGs and other genes encoding resistance to selective agents, such as heavy metals and disinfectants (Gillings, 2013; Gorecki et al., 2021; Rodríguez-Beltrán et al., 2021). Antibiotic resistance genes and metal resistance genes (MRGs) often co-occur on MGEs (Li et al., 2017; Wales & Davies, 2015). Plasmid mobilization is also of particular significance in general bacterial evolution, especially in terms of adaptation to changing environmental conditions, such as those induced by climate change (Dziewit et al., 2015; Gorecki et al., 2021). The presence of contaminants in the environment including antibiotics, quaternary ammonium compounds, and other biocides and metals (e.g., zinc and copper) can create co-selective pressures, whereby a single agent can select for resistance to multiple agents and other antimicrobials because of the physical co-location of the genes conferring resistance (Wales & Davies, 2015). There are many reports indicating the transfer of plasmid-encoded genes for resistance to clinically important antibiotics such as quinolones, carbapenems and colistin, in Escherichia coli and Klebsiella pneumoniae, other Enterobacteriaceae and Pseudomonas aeruginosa isolated from raw meat, farm animals (Liu et al., 2016), inpatients (Ma et al., 2022; Martínez-Martínez et al., 1998; Quan et al., 2023) and aquatic environments, raising serious concerns about its possible cross-environment dissemination (Zhu et al., 2020).
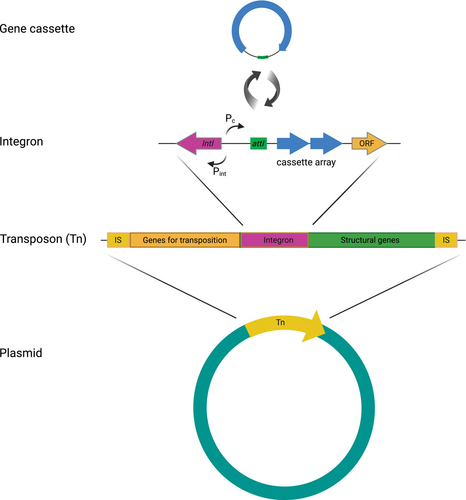
Integrons, which are often located on plasmids, also play an important role in the spread of multidrug resistance (Mazel, 2006). Integrons are DNA fragments equipped with gene cassettes that are capable of capturing a multitude of ARGs and other genes conferring resistance to antimicrobials, including disinfectants and heavy metals (Figure 1) (Gillings, 2014; Mazel, 2006). In Arctic environments, integrons have been implicated in the capture of genes encoding proteins thought to facilitate adaptation of bacteria to extreme environments (Makowska et al., 2020; Makowska-Zawierucha et al., 2022). However, data on the abundance of genes located in the structure of integrons in plasmidomes from Arctic environments are missing.
The widespread application of metagenomic sequencing has offered transformative new insights into the nature of environmental resistomes (Kirstahler et al., 2021; Norman et al., 2014; Stalder et al., 2019); however, typical approaches tend to neglect the plasmidome (Gillings, 2013; Martínez et al., 2017).
The aim of this study was to explore antibiotic resistance spread across human-affected and pristine environments in the high Arctic by comparing various aspects of the plasmidome across a gradient of pristine to anthropogenically affected environments in Spitsbergen—the largest island in Arctic Svalbard archipelago (European Arctic). The release of ca. 30 km3 of freshwater from glaciers and snow annually (Hagen et al., 2003), as well as the presence of polar stations and settlements, makes Svalbard a fitting study area for the investigation of links between climate induced shifts, microbial resistance and anthropogenic activity in the Arctic. We analyzed plasmidomes across distinct environments comprising: (i) surface glacial ice cores representing an environment with presumably minimal biological activity, relatively less affected by anthropogenic activities and likely hosting modern and ancient genetic material; (ii) rivers, fjords and aquatic sediments impacted by various levels of anthropogenic input; and (iii) treated and untreated wastewater outflows from settlements in Svalbard, representing specific human inputs. We apply metagenomic sequencing of the plasmidome fraction, which was accessed by first employing an enrichment step on generalized heterotrophic bacterial media and applying a plasmid-specific extraction protocol. We compared relative occurrences of stress-response genes in plasmidomes across the environments, including ARGs, biocide resistance genes (BRGs), MRGs and virulence genes (VGs). Specifically, we assessed the role of plasmids in the dissemination of AMR in Arctic environments over a range of anthropogenic impact. This study provides insight into natural baselines of AMR and how anthropogenic activities influence the contribution of plasmids and associated integrons to Arctic resistomes. Further, we found evidence that melting glaciers contribute plasmids carrying a diverse array of ARGs, BRGs and VGs to neighboring aquatic environments.
2 MATERIALS AND METHODS
2.1 Study sites and sampling
Samples were collected during three summer seasons (2020, 2021, 2022) from Spitsbergen, the largest island of the Svalbard Archipelago (Figure 2). In total, 20 samples were collected (Table S1). To simultaneously investigate the impact of the melting cryosphere and human activities on the resistome, sampling was conducted in contrasting systems along two gradients of anthropogenic influence: (i) the Longyearbyen city gradient (from glaciers, through the city with approximately 2400 inhabitants, to the fjord): a transect of glacial ice, river water and sediments, wastewater, and fjord waters and sediments, and (ii) the wastewater treatment plant (WWTP) outfall gradient in Ny-Ålesund, a permanent research settlement with a maximum summertime population of approximately 150 people in the northwest of the Svalbard archipelago. The Longyearbyen transect offers a gradient through the most populated, industrialized and touristed capital of Svalbard, spanning valley glaciers (Larsbreen and Longyearbreen) that feed the Longyearelva River, which crosses the city and flows into the fjord of Adventfjorden. Untreated wastewater from the city is released to Adventfjorden approximately 2-km offshore from Longyearbyen, via a sewerage effluent approximately 50–60 m below the water surface. For the Longyearbyen transect, sampling was carried out at (i) glaciers impacted by humans (Longyearbreen Glacier and Larsbreen Glacier located near Longyearbyen city, both visited by tourists, scientists and students from a local university) where ice cores were collected (in total eight ice cores), (ii) the Longyearelva river catchment (emanating from Longyearbreen and powered by streams from Larsbreen) where freshwater and sediments were collected (7 L of water, 1 kg of sediments), (iii) the seashore, where marine water and sediments were collected (7 L of water, 1 kg of sediments), and (iv) Longyearbyen's wastewater outflow (5 L of bottom water), then (v) crossing the Adventfjorden (5 L of bottom water) until reaching the Isfjorden system (5 L of bottom water)—the largest fjord on western Spitsbergen (at the border between Adventfjorden and Isfjorden).
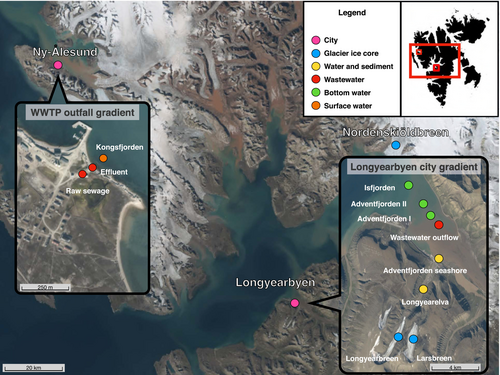
The water from the bottom of Adventfjorden (water collected directly above the bottom of the fjord) was collected from a rubber boat by using a 5-L-volume water sampler deployed to the bottom of the fjord using rope (60 m water depth). Ice cores were collected by using a Kovacs coring drill (Mark II, Ø = 7.5 cm). Each core was 40 cm in length. In the laboratory, the upper 5–10 cm of the ice core (so-called weathering crust) was cut and discarded to avoid windblown contamination and focusing on biological material stored in glacial ice. Although ice is considered largely impermeable, water on the surface of glaciers could percolate slowly inside the glacier through veins, incisions and channels. The flux of water into the below surface layers depends on the type of ice and meteorological conditions including rain (Fountain & Walder, 1998). For example, the Longyearbreen Glacier is a cold-type glacier with supraglacial (on ice) and englacial (in ice) channels, and incisions in the ice (Gulley et al., 2009; Sevestre et al., 2015), which could promote accumulation of pollutants in microconduits of glacial ice. The presence of water veins, the wetness of the ice, and finally the type of glacier could promote seeping of surface meltwater in specific areas of the glacier (Brown et al., 2017; Fountain & Walder, 1998), then stored contaminants already deposited on the ice surface. Therefore, we decided to drill cores in near-city and remote glaciers.
For the WWTP gradient in Ny-Ålesund, we collected samples from the WWTP outfall, Kongsfjord, north-west Svalbard (a small permanent settlement of international scientific and research stations) in order to compare the effects of WWTP and no-treatment plant on the marine resistome. (i) Raw sewage (2 L), (ii) effluent (2 L), and (iii) surface water from Kongsfjord (2 L), which is the effluent receiver, were collected. As a reference sample without direct anthropogenic impact, the marine terminating Nordenskiöldbreen Glacier was sampled (two ice cores). Nordenskiöldbreen is the least visited and the largest glacier among those investigated in this study.
The characteristics of the collected material and the location are presented in Table S1.
2.2 Accessing and extracting plasmid DNA
Due to challenges isolating plasmids from environmental samples, we employed an enrichment step prior to plasmid isolation. Water samples were filtered through 0.45-μm and then 0.2-μm cellulose nitrate filters (Sartorius Stedim): Adventfjorden seashore (2 L), Longyearelva (2 L), wastewater outflow in Longyearbyen (1 L), Adventfjorden I, II and Isfjorden, raw sewage, effluent and Kongsfjord as well as from Larsbreen, Longyearbreen (each 0.8 L, ice cores after thawing at 3°C) and Nordenskiöldbreen (3.3 L, ice core after thawing at 3°C). The filters were washed with sterile water and after centrifugation the pellet was inoculated onto 200 mL of R2A medium for samples from glaciers and Brain Heart Infusion (BHI) Broth for remaining samples and enriched for 48 h at 22°C on a shaker at 110 rpm according to the protocol of Gorecki et al. (2021).
After enrichment, plasmid DNA (pDNA) extraction was performed using a GeneJET Plasmid Midiprep Kit (ThermoFisher Scientific™). pDNA from sediment samples (Longyearelva, Adventfjorden seashore) was isolated from 5 g (wet weight) of each sample. Sediment samples were suspended in 50 mL of sterile water, vortexed for 30 min with sterile glass beads on a rotor at 120 rpm, then the suspension was centrifuged for 2 min and 5 mL was inoculated into 200 mL of BHI Broth for enrichment, and pDNA was extracted from the pellet as described above. To remove possible traces of genomic DNA, the precipitate was treated with plasmid-safe ATP-dependent DNase (Biosearch™ Technologies) according to the manufacturer's instructions. The pDNA was quantified using a Qubit 3 Fluorometer (ThermoFisher Scientific) according to the manufacturer's instructions.
2.3 Illumina high-throughput sequencing and sequence data analysis
Plasmidome sequencing libraries were prepared from 1 μg of input DNA fragmented to 200 bp using a Covaris E210 sonicator (Covaris, USA). DNA libraries were prepared using a NEBNext Ultra II DNA Library Prep Kit for Illumina (New England Biolabs, USA). DNA libraries were sequenced by Genomed SA company (Warsaw, Poland) on an Illumina NovaSeq 6000 system, using a paired-end read length of 2 × 150 bp with the Illumina NovaSeq 6000 reagent kits (Illumina, USA). The number of raw reads reached around 50 G base-pairs per library. Adapter sequences, primers and poly-A tails were removed using the Cutadapt version 4.0 (Martin, 2011). Error correction based on kmer method were conducted using Lighter version 1.1.2 (Song et al., 2014). Reads were assembled using metaplasmidSPAdes version 3.15.5 (Antipov et al., 2019) with default settings. Prodigal version 2.6.3 was used to predict protein-coding genes among assembled contigs. The NCBI Antimicrobial Resistance Gene Finder (AMRFinderPlus) version 3.11.11 (Feldgarden et al., 2021) and the accompanying database were used to find stress-response genes and VGs. Integrons were detected using Integron Finder version 1.5.1 (Néron et al., 2022). Gene cassettes located in the integrons were annotated using the INTEGRALL database (Moura et al., 2009). If the gene cassette structure (integron integrase nearby attC site/s) was not assembled, genes were considered as non-localized in integrons.
Heatmap visualization was conducted in R version 4.3.1 using ggplot2 version 3.4.2 (Wickham, 2009) and reshape2 version 1.4.4 (Wickham, 2007) packages and further graphically edited in Corel Draw 2021.
2.4 Statistical analysis
2.5 Direct DNA extraction and quantitative PCR for relative abundance calculations
Metagenomic DNA isolations were performed from replicates of the above samples (same volume of water samples and 1 g of sediment samples) using the DNeasy PowerSoil Pro Kits (Qiagen), according to manufacturer's instructions. Quality of the obtained DNA samples was assessed fluorometrically and by electrophoresis.
Gene quantities were normalized to the 16S rRNA gene copy numbers measured in whole community DNA using Droplet digital PCR (ddPCR) with primer sequences previously described by Maeda et al. (2003) and relative abundance values were expressed as percentages and calculated as follows: [(abundance/16S rRNA gene copy number) × 4 × 100%] with four being the average number of copies of the gene encoding 16S rRNA per bacterial cell, according to the ribosomal RNA database (Stalder et al., 2012). Each ddPCR was prepared in three technical replications. Reactions were conducted in a QX200 Droplet Digital PCR System (Bio-Rad) with QX200 ddPCR EvaGreen Supermix (Bio-Rad).
3 RESULTS
3.1 Stress-response genes
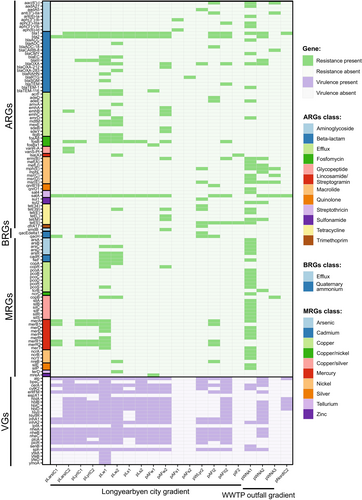
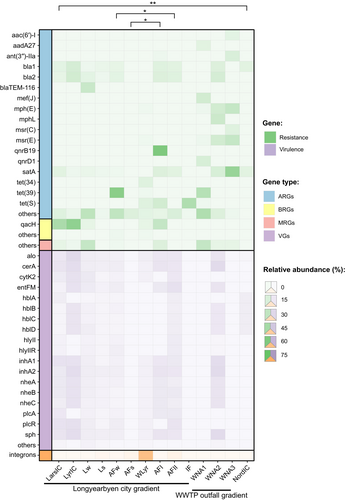
A total of 67 ARGs were identified in the plasmidome across all samples, with most conferring multidrug resistance (efflux pump) or resistance to β-lactams, aminoglycosides, macrolides, tetracyclines, fosfomycin, glycopeptides, quinolones, streptothricin, sulfonamides, lincosamides, streptogramins, or trimethoprim (Figure 3; Table S4). This analysis revealed the presence of three resistance mechanisms: antibiotic inactivation, antibiotic efflux and antibiotic target alteration/protection (Table S3). Furthermore, four ARGs in the plasmidome were shared across the greatest number of samples, that is, satA, bla1, bla2, and blaIII (Figure S1).
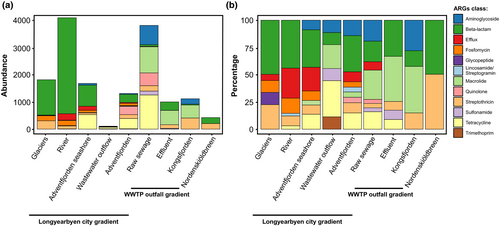
We found that both the diversity of ARGs and the number of classes of antibiotics to which the ARGs encoded resistance increased with increasing anthropogenic inputs (Figure 4a,b). For the Longyearbyen city gradient, in the glacial environment, resistance to β-lactams and streptothricin was found to be dominant (Figure 4a,b). In the wastewater outflow in Longyearbyen, resistance to tetracyclines and macrolides was found to be dominant, whilst in the Adventfjorden receiving wastewater, ARGs encoding resistance to β-lactams and aminoglycosides and tetracyclines were dominant (Figure 4b). In the WWTP outfall gradient, in effluent discharged into the fjord, ARGs encoding resistance to β-lactams were found, as well as ARGs encoding resistance to macrolides, streptothricin, sulfonamides, and tetracyclines. In the water receiving the effluent, resistance to macrolides and aminoglycosides was found to be dominant (Figure 4b).
A total of 41 MRGs were identified in the plasmidome, conferring resistance to 10 metals, with the dominant resistance categories being mercury, copper, silver, arsenic, and nickel (Figure 3; Table S4). Moreover, merA conferring resistance to mercury was shared in the largest number of samples (Figure S1). Our data indicate that water from the river as well as raw sewage environments contained the greatest diversity of MRGs (Figure 5a,b). No MRGs was detected in wastewater outflow or Nordenskiöldbreen ice core. In addition, a total of three BRGs that encode resistance to quaternary ammonium compounds (qacH, qacEdelta1) and smdB encoding a multidrug efflux pump ABC were identified in the plasmidome (Figure 3). Furthermore, qacH was shared among the largest number of environments.
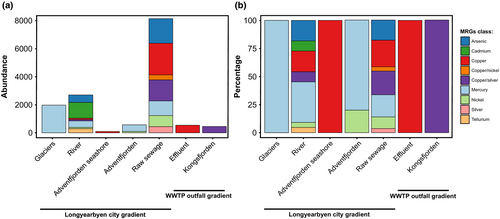
The relative abundance of stress-response genes varied among sampling sites. The relative abundance of ARGs ranged from 0.1% for lso(A) gene in water of Longyearelva, blaTEM-116 in water of Adventfjorden seashore, bla1, msrC, ermD, aac(6′)-I in raw sewage from WWTP and ermB in effluent from WWTP in Ny-Ålesund to 42% for the qnrB19 gene in bottom water of Adventfjorden after the discharge of wastewater (Figure 6; Table S2). The relative abundance of MRGs ranged from 0.1% for copA and copB to 4.3% for cadR in water of Longyearelva and tcrB in effluent from WWTP in Ny-Ålesund. In contrast, the relative abundance of BRGs ranged from 1.3% for qacEdelta1 in wastewater outflow in Longyearbyen to 36.5% for qacH in Longyearbreen Glacier. There was a significant increase of plasmid-carried stress-response genes relative abundance in bottom water of Adventfjorden after the discharge of wastewater (AFI, AFII) compared to water and sediment of Adventfjorden seashore (AFw, AFs) (p < .05). In addition, Figure 6 shows significant differences in stress-response genes between Nordenskiöldbreen and Longyearbreen glaciers (p < .001) with lower number of detected genes on Nordenskiöldbreen, which is more remote and less visited by tourists.
3.2 Virulence genes in the plasmidomes
A total of 26 VGs were identified across the plasmidomes. The production of exotoxins, including enterotoxins important for bacterial pathogenesis, was the dominant virulence mechanism encountered in the data (Figure 3; Table S3). Furthermore, 18 VGs encoding exotoxins that damage the host cell membrane by forming pores or hydrolyzing cell membrane lipids were shared in the largest number of samples (Figure S1). No VGs were found in the plasmidome isolated from the Adventfjorden sediment, Isfjorden bottom water and water receiving effluent from the WWTP in Ny-Ålesund (Figure 3).
The relative abundance of VGs varied between sampling sites; ranging from 0.1% for cerA, cytK2, inhA2 and sph in raw sewage from WWTP in Ny-Ålesund to 16.8% for alo in Longyearbreen Glacier (Figure 6; Table S2). There was a significant increase in the relative abundance of VGs in bottom water of Adventfjorden receiving wastewater discharge (AFII) compared to water of Adventfjorden seashore (p < .05). In addition, significant differences in VGs abundance were found between Nordenskiöldbreen and Longyearbreen glaciers (p < .001), with the lower abundance on the reference glacier, Nordenskiöldbreen.
3.3 Integrons associated with the plasmids
Integrons were identified in the plasmid extracts at all sites, except for the glacial ice core samples. The relative abundance of integrons varied between sampling sites and ranged from 0.03% in raw sewage from WWTP in Ny-Ålesund, to 40.3% in wastewater outflow in Longyearbyen (Figure 6; Table S5). Among all identified integrons, ARG cassettes were found in only one integron in the wastewater outflow in Adventfjorden (Table S6). In the variable region of this integron, gene cassette array dfrA17-ant(3′)-IIa confers resistance to trimethoprim and streptomycin. The remaining gene cassettes largely determine metabolic and adaptive functions (Table S6). The variable region of integrons also included other genes responsible for the production of membrane transporters and forming efflux pumps, IS110 family IS transposase protein, transposases involved in the transposition mechanism, and even genes for bacterial abortive infection (Abi) systems encoding phage resistance proteins that limit viral replication. In addition, 62.2% of all gene cassettes were hypothetical proteins with an unknown function.
3.4 Relationship between stress-response genes, virulence genes and integrons in an environment with distinct anthropogenic inputs
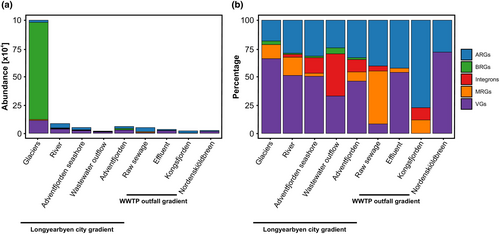
We found that the abundance and frequency with which VGs were detected was the highest within the Longyearbyen city transect and decreased as anthropogenic inputs increased through the Longyearelva River and Adventfjorden seashore (Figure 7a,b). Correspondingly, VGs were found to be higher in the effluent from WWTP in Ny-Ålesund and in the Adventfjorden water downstream of the wastewater outflow discharge than in the raw sewage (Figure 7a,b). The frequency of detections of ARGs increased in river water as well as in the effluent from WWTP in Ny-Ålesund and the fjord water receiving effluent in both Longyearbyen and Ny-Ålesund (Figure 7b), whilst the detection of integrons in Kongsfjorden was most frequent in the water receiving effluent from WWTP in Ny-Ålesund (Figure 7b). The highest frequency of detections of MRGs was found in raw sewage from WWTP in Ny-Ålesund. In contrast, BRGs dominated in the wastewater outflow in Longyearbyen.
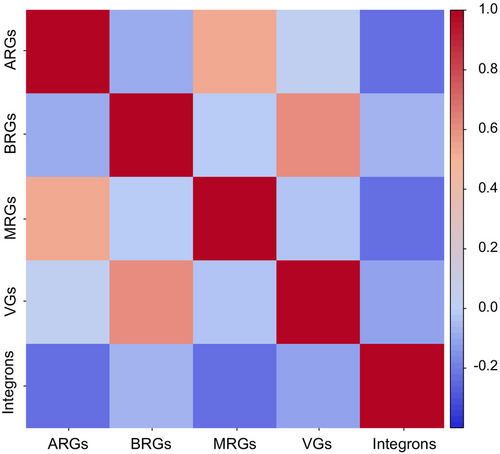
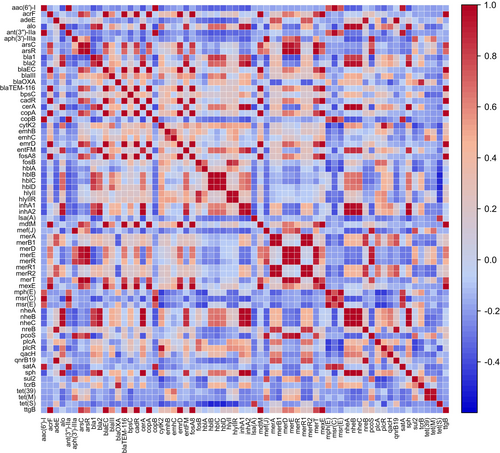
The relative abundance of ARGs strongly correlated with MRGs, but not with integrons, whilst VGs correlated with BRGs (Figure 8). The correlations among individual genes that were found in more than one sampling site is shown in Figure 9, which broadly shows that ARGs encoding resistance to β-lactams and VGs encoding hemolysins BL (Hbl), non-hemolytic enterotoxins (Nhe), hemolysins II (HlyII) and InhA metalloproteases co-occur, whilst macrolide resistance genes and MRGs conferring resistance to mercury do not correlate with the above VGs.
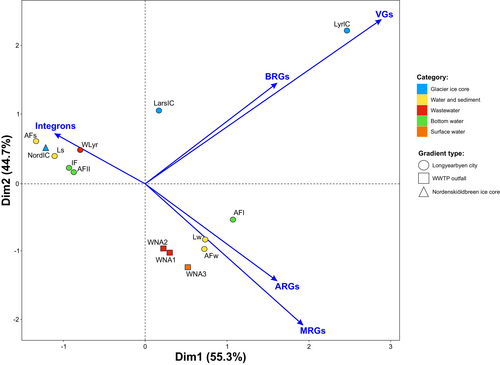
Principal component analysis along vector projection revealed that Larsbreen and Longyearbreen glaciers were characterized by plasmidomes that were dominant in BRGs, which were also correlated with VGs (Figure 10). In samples from WWTP in Ny-Ålesund (WNA1, WNA2) and surface water of Kongsfjorden (WNA3) as well as in water of Longyearelva (Lw), water of Adventfjorden seashore (AFw), bottom water of Adventfjorden downstream the discharge of wastewater (AFI), dominated by ARGs, which are correlated with the presence of MRGs (Figure 10). Integrons dominate in the samples from environments with different levels of anthropogenic input.
4 DISCUSSION
Numerous studies indicate the widespread distribution of ARGs in soils (Torres-Cortés et al., 2011), surface waters (Martínez, 2008; Pruden et al., 2012), WWTPs (Mao et al., 2015) and even remote, cold and rapidly changing polar regions (Makowska et al., 2020; Makowska-Zawierucha et al., 2022). Whilst cultivation-independent metagenomics of environmental samples provides information on the environmental resistome, it is difficult to elucidate the specific role of MGEs. Here, we explored a plasmidome from various Arctic samples with varying levels of anthropogenic input, by applying enrichment culturing plasmid isolation and high-throughput sequencing. Mapping the occurrence of plasmid-encoded genes in Arctic ecosystems can provide vital information toward understanding sources, sinks, and pathways for ARG spread and correspondingly informing appropriate efforts toward mitigating the spread of antibiotic resistance (WHO, 2021). Although the Arctic is generally considered to be a receiver of pollutants from lower latitudes, we demonstrate here that glaciers themselves are a repository of various genes associated with AMR and virulence which are delivered to the marine ecosystems. Considering that exported genetic material could be incorporated into adjacent and distant environments through ocean currents (Pittino et al., 2023; Yang et al., 2021), this study further implicates glaciers may be an important contributor to the marine resistome. Although this phenomenon is not presently recognized, the Gulf Stream and East Greenland Current could disseminate of waters carrying microbial resistance genes to Arctic regions and from the Arctic to lower latitudes. The emergence and rapid spread of ARGs poses a serious threat to public health, which, further aided by climate change, may increase the pool of ARGs available to pathogens (Martínez, 2008; McCann et al., 2019). We found 12 main types of ARGs in the plasmidome—mainly comprised of β-lactam, multidrug, aminoglycoside, macrolide and tetracycline resistance genes. Similar results have been reported in previous studies on metagenomes from Kongsfjorden soil cores (McCann et al., 2019), likely these mechanisms are plasmid encoded. Here, we found that the most common ARGs were the bla1 and bla2 genes, which were detected on plasmids across a variety of sampled environments regardless of the degree of anthropogenic input. Importantly, the bla2 gene is a metallo-β-lactamase that catalyzes the hydrolysis of a wide range of β-lactam drugs, including carbapenems, which are considered the antibiotics of last resort in the treatment of the most serious infections caused by Gram-negative bacteria (Naquin et al., 2017). Mogrovejo et al. (2020) found a high frequency of resistance to β-lactam and carbapenems in strains grown from Greenland and the Svalbard Archipelago. The relative abundance and variability of the ARGs significantly increased along the gradients from glaciers to wastewater outflow in the fjord, suggesting that human activity strongly influences the gene pool carried by the plasmidome, which in turn shapes the Arctic environmental resistome, corroborating other studies (McCann et al., 2019; Sajjad et al., 2020). Consequently, we suggest that the delivery of ARGs from glaciers and wastewater to marine ecosystems both act to increase the global reservoir of ARGs.
Among the 10 identified subtypes of MRGs, genes of the mer operon, which are involved in regulation, mercury (Hg) binding, and organomercury degradation, were found to be dominant in the glacial environments. Mercury—one of the most toxic heavy metals, causing changes in the structure of proteins leading to loss of function (Boyd & Barkay, 2012)—and its methylated forms are known to have accumulated in various environments, including polar and high mountain regions (Dietz et al., 2009; Douglas et al., 2012; Sun et al., 2017). High concentrations of heavy metals have been detected on glaciers compared to glacier adjacent sites, due to atmospheric fallout (Łokas et al., 2016). Previous studies reporting metal resistance in cultured polar microorganisms include those isolated from cryoconite holes on glaciers (Pittino et al., 2023) and snow from the high Arctic, where 31% of the culturable bacteria are Hg-resistant, compared to less than 2% in nearby freshwater and brine samples (Møller et al., 2014). Indeed Larose et al. (2013) found that the concentration of Hg in snow correlates with the number of copies of the Hg resistance gene among bacterial communities. Furthermore, it was found that the merA gene is most often located on plasmids (Møller et al., 2014).
The anthropogenic contribution to Hg occurrence in the environment has been found to be dominant (over 92%), which suggests that the Arctic cannot be considered a region that is shielded from the impact of human activity (Dietz et al., 2009; Łokas et al., 2016). The mechanism of action of the merA gene, which encodes mercuric reductase, is to catalyze the reduction of Hg2+ to volatile metallic mercury Hg0 (Boyd & Barkay, 2012). Previous studies have found that the percentage of Hg-resistant strains in Arctic coastal waters can explain most of the Hg0 in this aquatic environment (Poulain et al., 2007). Moreover, the presence of genes encoding resistance to copper, silver, arsenic, and nickel can be explained by high concentrations of the respective metals in the Arctic environments (Hauptmann et al., 2017). Our results suggest that the pressure caused by the presence of metals in Arctic environments contributes not only to the maintenance of metal resistance but also to the emergence and spread of ARGs that co-occur on the same MGEs (Baker-Austin et al., 2006; Perez et al., 2020). Moreover, the strong relationship between the occurrence of MRGs and ARGs suggests that MRGs may be a marker of broader AMR in various environments, including polar ecosystems.
Virulence genes allow infection of hosts by pathogenic bacteria. Virulence genes can be located on the chromosome or MGEs (e.g., plasmids or transposons) and encode activities such as adhesion, invasion, attachment, iron acquisition, motility, and toxin activity (Wu et al., 2008). Currently, there is little information on the occurrence of VGs in the Arctic. Previous studies revealed the presence of VGs in Arctic permafrost that also co-occurred with ARGs on MGEs (Kim et al., 2022). Our results indicate that the production of exotoxins, including enterotoxins, is the dominant virulence mechanism in human-affected Arctic environments.
We found that the frequency of detections of VGs decreases along the gradient from the glacial environment, through the Longyearelva River, which flows into the Adventfjorden, whilst the relative abundance of VGs significantly increases with the level of anthropogenic input, as exemplified by the discharge of wastewater into Adventfjorden and in WWTP effluent. The significant differences between VGs in glacial and non-glacial environments may point toward the physical features of Arctic environments such as depositional processes (e.g., accumulation versus dilution between glaciers and fjords respectively) shaping VGs diversity and abundance. Our study indicates a potential role of glaciers in releasing VGs: we demonstrated a high abundance of virulence factors in environments characterized by low anthropogenic input (e.g., glaciers), which has been documented by earlier reports (Kim et al., 2022), corroborating evidence for a strong pressure of viruses on heterotrophic bacteria on glaciers (Bellas et al., 2020). Viruses present in the glacial environment may affect the pathogenicity of bacterial communities through the transfer of virus-encoded VGs (Bi et al., 2023). Furthermore, bacteria released from melting glaciers may harbor VGs homologues and are unlikely to be opportunistic human pathogens (Søborg et al., 2016). These genes may be maintained for benefit, or for co-occurrence with other genes conferring benefits for bacterial survival outside the host environment (Brown et al., 2012).
Glacial run-off and iceberg calving may deliver VGs to bacterial communities in downstream environments, which may potentially enhance virulence traits for opportunistic pathogenic bacteria (Sajjad et al., 2020; Søborg et al., 2016).
Integrons are carriers of gene cassettes encoding various functions and are most often located in transposons and on plasmids (Gillings, 2014). However, in many human-affected environments, integrons are involved in the spread of multidrug resistance (Gillings, 2013, 2014). Makowska et al. (2020) and Makowska-Zawierucha et al. (2022) have shown the presence of strains with class 1 integrons isolated from both glacial environments in Svalbard (in cryoconite on the surface of glaciers and glacial ice) and seawater of fjords, including wastewater outflow. Previous research showed that in the variable regions of integrons in culturable bacteria, the occurrence of genes determining different functions, including antibiotic resistance, virulence and physiology was noted, highlighting the crucial role of these genes in adaptation of bacteria to stress (osmotic, light, temperature) in Arctic environments (Makowska-Zawierucha et al., 2022).
In a previous metagenomic study, McCann et al. (2019) found intI1 in high Arctic soil ecosystems, indicating the development of antibiotic resistance in remote terrestrial ecosystems. Whilst other studies have investigated ARGs and ARB in snow and ice of Svalbard and other cold regions, no data on integrons were provided (Segawa et al., 2013). In our study, we found that the integrons located in the Arctic plasmidome contain mainly adaptive genes and genes for hypothetical proteins with unknown functions. Resistance genes have only been identified in wastewater, which points toward a relationship between ARGs and anthropogenic input. Our results indicate that ARGs, BRGs, MRGs, and VGs are in the plasmid, but outside of the integron structure. The presence of integrons with no typical ARGs inserted into the variable region may indicate that ARG cassettes are excised from the structure in the absence of antibiotic selective pressure, whilst representing elements, which could acquire and create new gene combinations (Stokes et al., 2006). The absence of ARGs in the structure of integrons in the plasmidome may also indicate unique features of the microbial community in this extreme environment.
Our findings show that plasmid-encoded antibiotic resistance is widespread in the Arctic environment, where selection pressure for antibiotics is presumed to be low. Plasmid-encoded antibiotic resistance may be derived from both anthropogenic sources as well as the release of ancient variants of genes from the ablating cryosphere. Moreover, the presence of MRGs on plasmids combined with the prevalence of metals in the environment could potentially facilitate the spread of resistance by HGT. A strong correlation between MRGs and ARGs confirms their co-occurrence and can create the opportunity for co-selective pressure. Moreover, we showed differences in the relative abundance of ARGs, BRGs, MRGs and VGs in environments with varying human impacts. In the glacial environment, VGs and BRGs predominate, which suggests that glacial ice may be a repository of pathogenic strains with associated genes on the plasmids and may accumulate contaminants currently considered biotic pollutants, which mix with water from melting glaciers and may disseminate into the environment (Makowska-Zawierucha et al., 2022).
5 CONCLUSIONS
We examined the cultivable plasmidome across a diverse range of Arctic environments and found significant differences in stress-response gene and VG diversity and abundance across gradients in anthropogenic activity. Both stress-response genes and VGs were found to related with plasmids in samples from glacial environments, rivers, municipal sewage and the marine environment. We found that ARGs and MRGs are dominant in aquatic environments whilst VGs and BRGs are dominant in glacial ice, suggesting that the resistance and virulence in bacteria may be shaped by differences in the physical environment, accumulation of pollutants or nutrient availability. We found that whilst the level of antibiotic and metal resistance varies between ecosystems in Arctic, all sources (glaciers, wastewater) from terrestrial ecosystems may increase the aquatic resistome gene pool by delivery to downstream environments and mixing in fjords and ocean basins. Considering the increasing meltwater export from Svalbard glaciers (total mass balance −8 ± 6 Gt a−1), glacier surface runoff reaches 25 ± 5 km3 a−1, and freshwater flux from calving accounts for 4 ± 1 km3 a−1 (Hagen et al., 2003; Schuler et al., 2020), meltwater containing both ARGs and VGs may affect function and processes of downstream ecosystems. The widespread occurrence of genes determining physiological functions and genes for hypothetical proteins with unknown functions in the variable regions of integrons, highlights a central role for these genes in adaptation to Arctic environments but also presents a risk of developing new resistance phenotypes. We did not find typical ARGs within the integron cassettes, suggesting that integrons may not play as important a role in the spread of ARGs in Arctic as previously thought. Nevertheless, as genetic material from different sources including anthropogenic inputs mixes downstream and in fjords, it may be incorporated into the wider aquatic environment.
AUTHOR CONTRIBUTIONS
Nicoletta Makowska-Zawierucha: Conceptualization; data curation; formal analysis; funding acquisition; investigation; methodology; project administration; resources; supervision; validation; visualization; writing – original draft; writing – review and editing. Artur Trzebny: Data curation; formal analysis; methodology; visualization; writing – review and editing. Krzysztof Zawierucha: Investigation; methodology; writing – original draft; writing – review and editing. Vineeth Manthapuri: Formal analysis; visualization; writing – review and editing. James A. Bradley: Investigation; writing – review and editing. Amy Pruden: Writing – original draft; writing – review and editing.
ACKNOWLEDGMENTS
This study was supported by grant no. 2020/36/C/NZ9/00221 from the National Science Centre, Poland. The project is listed in the Research in Svalbard database under RiS ID 11563. Authors thanks three anonymous reviewers for their valuable comments on the manuscript. We thank the staff of Kings Bay AS for assistance with wastewater sampling in Ny-Ålesund. We gratefully acknowledge Poland's high-performance computing infrastructure PLGrid (HPC Centers: ACK Cyfronet AGH) for providing computer facilities and support within computational grant no. PLG/2023/016110. JAB was supported by Natural Environment Research Council (NERC) grant NE/T010967/1 and the Centre national de la recherche scientifique (CNRS) Chaires de Professeur Junior (CPJ). The Virginia Tech team was additionally supported by US National Science Foundation Awards OAC 2004751, OISE 1545756, NRT 2125798, and ECCS 2025151. We would like to thank Piotr Bałazy, Maciej Chełchowski, Dariusz Ignatiuk and Piotr Rozwalak for field support.
CONFLICT OF INTEREST STATEMENT
The authors declare no competing interest.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are openly available in NCBI under BioProject accession number PRJNA983705 at https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA983705.




