Allopatric origin of sympatric whitefish morphs with insights on the genetic basis of their reproductive isolation
Abstract
The European whitefish (Coregonus lavaretus) species complex is a classic example of recent adaptive radiation. Here, we examine a whitefish population introduced to northern Finnish Lake Tsahkal in the late 1960s, where three divergent morphs (viz. littoral, pelagic, and profundal feeders) were found 10 generations after. Using demographic modeling based on genomic data, we show that whitefish morphs evolved during a phase of strict isolation, refuting a rapid sympatric divergence scenario. The lake is now an artificial hybrid zone between morphs originated in allopatry. Despite their current syntopy, clear genetic differentiation remains between two of the three morphs. Using admixture mapping, we identify five SNPs associated with gonad weight variation, a proxy for sexual maturity and spawning time. We suggest that ecological adaptations in spawning time evolved in allopatry are currently maintaining partial reproductive isolation in the absence of other barriers to gene flow.
The pace at which evolution and speciation occur, and the mechanisms determining their tempo, has intrigued evolutionary biologists for a long time (Simpson 1944; Bush et al. 1977; Kornfield 1978), and still continue today (Schluter 2000; Nosil 2012; Lescak et al. 2015; Momigliano et al. 2017; Salzburger 2018; Skúlason et al. 2019). Concerns over the organisms’ ability to adapt to environmental changes (e.g., Gienapp et al. 2008; Merilä and Hendry 2014), and the realization that adaptation can be very rapid (Thomas et al. 2017; Matthews et al. 2018; Marques et al. 2019b), have sparked new interest toward the rates of evolution. Adaptation from standing genetic variation can be fast, especially if genetic diversity has been augmented by historical gene flow (Häkli et al. 2018; Salzburger 2018; Jacobs et al. 2019; Marques et al. 2019a). One example is the evolution of freshwater-adapted three-spined sticklebacks (Gasterosteus aculeatus) colonizing ponds that were formed during uplift caused by the 1964 Great Alaska Earthquake (Lescak et al. 2015). Similarly rapid adaptation has been observed for sockeye salmon and Darwin's finches when colonizing new environments (Hendry et al. 2000; Lamichhaney et al. 2018). In all these examples, broad ecological opportunities in novel environments likely facilitated rapid phenotypic and genetic differentiation.
Coregonid fishes have undergone extensive recent adaptive radiations but intermittent gene flow among morphs and species is common (Østbye et al. 2005; Bernatchez et al. 2010; Hudson et al. 2011; Vonlanthen et al. 2012). European whitefish (Coregonus lavaretus) experienced frequent adaptive radiations throughout their distribution (Svärdson 1979; Østbye et al. 2005; Vonlanthen et al. 2012). Facing different ecological opportunities, C. lavaretus evolved partially reproductively isolated, co-existing morphs using different niches (Kahilainen and Østbye 2006; Siwertsson et al. 2010; Præbel et al. 2013). These morphs differ in several phenotypic traits (Harrod et al. 2010; Præbel et al. 2013), such as body size and number of gill rakers, both heritable traits related to feeding ecology (Kahilainen et al. 2011; Häkli et al. 2018). Although rapid whitefish differentiation has been documented in both Nearctic and Palearctic regions (Bernatchez et al. 2010; Hudson et al. 2011), less is known about the genetic basis of their rapid phenotypic differentiation (but see Vonlanthen et al. 2012; Jacobs et al. 2019; Rougeux et al. 2019a).
During the 1960s, the Finnish Fisheries authorities facilitated introductions of whitefish in several high-altitude lakes including Lake Tsahkal, where the species did not previously occur. Sampling in Lake Tsahkal in 2011 revealed three different morphs inhabiting littoral, pelagic, and profundal habitats (Kahilainen et al. 2011; Præbel et al. 2013). There is evidence of both allopatric and sympatric divergence of whitefish morphs (Præbel et al. 2013; Salisbury and Ruzzante 2022), and genetic diversity fueled by historical admixture can facilitate rapid phenotypic divergence when ecological opportunity arises (Jacobs et al. 2019; Frei et al. 2022). It is possible that the whitefish introduced to Lake Tsahkal came from a single ancestral population and gave rise to different morphs in about 50 years (≈10 generations). The alternative hypothesis is that two or three morphs were simultaneously introduced in the lake. In such case, the whitefish of Lake Tsahkal would provide a rare opportunity to study the ecological and genetic underpinnings of reproductive barriers following secondary contact.
Lake Tsahkal provides also an ideal system to study the genetic basis of phenotypic traits defining the whitefish morphs via admixture mapping (Gompert et al. 2017). To date, few studies have provided insights on the genetic basis of whitefish phenotypic traits (Vonlanthen et al. 2009; Gagnaire et al. 2013a,b; Feulner and Seehausen 2019; Jacobs et al. 2019). These studies focus mainly on trophic traits, whereas less attention has been paid on traits associated with annual timing of reproduction. Timing of reproduction can act as a reproductive barrier among diverging populations (Hendry and Day 2005), and it is well-known that whitefish morphs have differences in their spawning time and location (e.g., Svärdson 1979; Vonlanthen et al. 2009; Kahilainen et al. 2014; Bitz-Thorsen et al. 2020).
Here, using a demographic modeling framework, we test two possible divergence scenarios of the Lake Tsahkal whitefish morphs: sympatric divergence and allopatric divergence followed by secondary contact. Furthermore, we investigate the genetic architecture of 22 morphological traits, including gonad weight variation, a proxy for spawning time that is a trait expected to be under ecological selection that could lead to reproductive isolation.
Methods
STUDY SYSTEM AND DATA COLLECTION
Lake Tsahkal is located in the tree line of northern Finnish Lapland (69°01ʹN, 20°50ʹE, 559 m a.s.l.). The lake is oligotrophic (totP 5 μg/L, totN 140 μg/L), deep (max 35 m, mean 9 m), clearwater (compensation depth 7.5 m), and has equal habitat distribution (41% pelagic/profundal, 59% littoral; Hayden et al. 2014). Introduced whitefish in the 1960s had broad ecological opportunity as they are more efficient in zooplanktivory and benthivory than two native species in the lake: the brown trout (Salmo trutta) and burbot (Lota lota) (Siwertsson et al. 2010; Hayden et al. 2014). Reproductive isolation among whitefish morphs could arise via differences in resource use causing differences in spawning times and places (Kahilainen et al. 2014; Taylor and Friesen 2017; Thibert-Plante et al. 2020).
Littoral, pelagic, and profundal zones were identified in the lake (Hayden et al. 2014) (Fig. 1a) and subsequently whitefish were collected from these habitats with gill net series in August 3−17, 2011 (Kahilainen et al. 2011). Caught fish were transported to field laboratory and labeled and frozen (−20°C) for later measurements of fish size, gill raker count, resource use metrics, and morphological traits (for details, see Table S1, Method S1, and Figs. S1, S2, S4). All caught fish were assigned to three different morphs according to their gill raker appearance, head, and body shape (Method S1; Kahilainen and Østbye 2006).
A total of 61 individuals (17 littoral, 22 pelagic, and 22 profundal fish) were sequenced using 2b-RAD sequencing approach (Wang et al. 2012) to obtain 36 bp single-end fragments with a mean coverage 30.2× (13.7−44.0; Table S1) by Illumina HiSeq 4000 at BGI, Hong Kong. DNA extraction and 2b-RAD library preparation followed exactly the protocols detailed in Momigliano et al. (2018, 2021).
POPULATION GENETIC ANALYSES
Raw reads were demultiplexed and PCR duplicates removed as per Momigliano et al. (2018). Reads were aligned to a chromosome-level assembly of the alpine European whitefish (Coregonus spp.) publicly available at https://www.ebi.ac.uk/ena/data/view/GCA_9021750 (De-Kayne et al. 2020), using Bowtie2 (Langmead and Salzberg 2012). SAM files were converted to BAM files and indexed using SAMtools (Li and Durbin 2009).
Genotype likelihoods were estimated from BAM files using ANGSD (Korneliussen et al. 2014), retaining only biallelic loci with ≤25% missing data and bases with mapping quality and Phred scores >20. We performed a principal component analysis (PCA) using PCAngsd (Meisner and Albrechtsen 2018) retaining variants with minimum minor allele frequency (MAF) of 0.02. Individual ancestries were inferred using NGSadmix (Skotte et al. 2012) based on genotype likelihoods, assuming one to three ancestral populations. Absolute divergence (dXY) (Nei 1987) and net nucleotide divergence (Da) (Nei 1987) were estimated based on nonadmixed individuals identified by NGSadmix using scripts from Momigliano et al. (2021).
DEMOGRAPHIC MODELING
We compared demographic models using the software package moments (Jouganous et al. 2017) based on diffusion approximations of the allele frequency spectrum (SFS). Ten demographic models were tested using nonadmixed littoral and pelagic individuals, as the ancestral pelagic and profundal morphs were not genetically distinct (see Results) and we had more pelagic (n = 8) than profundal (n = 5) nonadmixed individuals. We first defined five competing gene flow scenarios: strict isolation (SI), isolation with migration (IM), secondary contact (SC), ancient migration (AM), and a two epochs model (2EP) assuming heterogeneous migration rates through time (Roux et al. 2016; Momigliano et al. 2021). Because unaccounted changes in Ne can bias model choice and parameter estimation (Momigliano et al. 2021), we defined five additional models accounting for a change in Ne (increase or decrease) in both daughter populations at time T2, based on the above simple models: SI_NeC, AM_NeC, IM_NeC, SC_NeC, and 2EP_NeC; NeC stands for “Ne Change.” The 10 tested models are visualized in Figure S6. To account for possible effects of linkage in the 2b-RAD data, the best fitting model was chosen using Likelihood Ratio Test (LRT) as outlined in Coffman et al. (2016).
All analyses were performed based on the unfolded two-dimensional SFS (2D-SFS) derived from the sites shared between pairwise morphs. The programs ANGSD and Moments and custom R and python scripts adopted from Momigliano et al. (2021) and Fang et al. (2021) were used in the analyses. Detailed methods are given in Method S1.
ADMIXTURE MAPPING
Admixture mapping of phenotypic traits was performed with two genome-wide association (GWA) approaches: GEMMA (version 0.98), a genome-wide efficient mixed model association approach (Zhou and Stephens 2014), and LDna-EMMAX, a linkage disequilibrium (LD) clustering-based approach for association mapping (Kang et al. 2010; Kemppainen et al. 2015; Li et al. 2018; Fang et al. 2021). The same loci obtained from the filtering steps above were used in ANGSD for GWAs, except that we increased the MAF threshold to 0.1 (-minMaf 0.1), resulting in 12,212 loci. Genotype likelihood data were used in LDna-EMMAX, whereas called genotypes derived from ANGSD (-doVcf 1) were fed to GEMMA.
Univariate linear mixed models were conducted for each phenotypic trait in GEMMA, accounting for relatedness between individuals by supplying a relatedness matrix as a covariate. In LDna-EMMAX, we used LD network analyses (LDna; Kemppainen et al. 2015) to identify correlated clusters of single-nucleotide polymorphisms (SNPs), followed by GWA by fitting a multilocus mixed model accounting for relatedness as in Li et al. (2018) and Fang et al. (2021). Assuming a linear relationship between trait and body size, phenotypic measurements were corrected to mean total length (22.4 cm) before GWAs (Method S1). Significance tests of the two GWA approaches were performed based on Wald and permutation tests (10,000 permutations), respectively.
We corrected for multiple testing using the false discovery rate (FDR) approach of Benjamini and Hochberg (1995) and Benjamini and Yekutieli (2001) with an FDR threshold of 0.1. Specifically, Q-values (the FDR at which a test is significant) were calculated from P-values using the R-function p.dajust and we performed this in two ways. First, for GEMMA, the P-values derived from Wald-tests were corrected for genome-wide 12,212 SNPs; for LDna, we simply adopted P-values estimated by 10,000 permutations. Second, in a more stringent correction, the number of traits (22) was corrected for GEMMA (12,212 × 22 = 268,664 independent tests) and LDna-EMMAX (22 independent tests). To visualize phenotype-genotype correlation, we performed with PCAngsd a PCA based on the genotype likelihoods of the loci associated with gonad weight variation, and tested correlations between the first principal component (PC1) and adjusted gonad weight of individuals. This test was also conducted for genome-wide neutral SNPs for comparison. Details and pipelines used to perform GWAs are provided in Method S1.
Results
POPULATION STRUCTURE AND DEMOGRAPHIC HISTORY
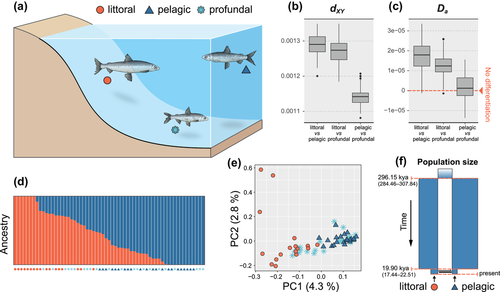
Littoral and pelagic whitefish morphs differed significantly in body size, gill raker count, morphology, diet, and parasites, whereas the profundal morph was intermediate (Fig. S4). Stable isotopes and mercury indicated clear differentiation of all three morphs in their respective habitats (Fig. S4). We obtained genotype likelihoods of 15,615 loci across 61 individuals. NGSadmix analyses revealed two ancestral components (K = 2), and a clear genetic differentiation between littoral and pelagic/profundal morphs in analysis of nonadmixed individuals (Fig. 1b–d). Overall divergence between nonadmixed pelagic and profundal morphs was low: dXY = 1.14 × 10−3 (95% confidence interval [CI]: 1.08 × 10−3 to 1.21 × 10−3; Fig. 1b) and Da = 1.81 × 10−6 (95% CI: −1.37 × 10−5 to 1.56 × 10−5; Fig. 1c). Out of 61 individuals analyzed, 41 were genetically admixed (i.e., having >3% ancestry from a secondary cluster; Fig. 1d). PCA supports these inferences, showing no differentiation of pelagic and profundal morphs, and admixture between littoral and pelagic morphs (Fig. 1e).
The demographic analyses supported secondary contact with changes in Ne (SC_NeC; Figs. 1f, S7, S8) as the best-fit model. Littoral and pelagic morphs diverged 296.15 thousand years ago (kya; 95% CI: 284.46−307.84 kya, i.e., 56.89−61.57 thousand generations), and experienced a secondary contact following the end of the last glaciation 19.9 kya (95% CI: 17.44−22.51 kya, 3.49−4.50 thousand generations) with decreases in Ne for both morphs after the Last Glacial Maximum (LGM; Fig. 1f). For a summary of scaled parameters, see Table S2.
ADMIXTURE MAPPING OF GONADAL SIZE VARIATION
The two GWA analyses collectively revealed five SNPs (Chromosome [Chr] 22: 43,111,579, Chr28: 9,855,214, Chr28 9,763,679, Chr32 37,564,809, and Chr36: 8,058,628) associated with gonad weight variation in the 41 admixed individuals (Fig. 2). Out of these, two SNPs in Chr22: 43,111,579 and Chr36: 8,058,628 were in strong LD (r2 > 0.9) and clustered in LDna (the first step in LDna-EMMAX analyses).
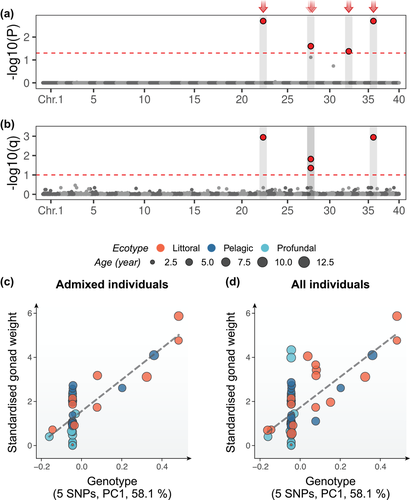
The genotypes of the five SNPs explained 63% of the variation in gonad weight among admixed individuals (P < 6.5 × 10−10; Fig. 2c), 44% among all collected individuals (P < 7.5 × 10−9; Fig. 2d), 44% among all female individuals (P < 2.3 × 10−5, Fig. S8), and 26% among all male individuals (P < 6.5 × 10−3; Fig. S8). The individual SNPs above explained 41%−51% of the variation in gonad weight among admixed individuals (Fig. S9). However, the genome-wide neutral loci did not explain any variation in gonadal weight as shown by nonsignificant phenotype-genotype correlation (Fig. S8). When correcting for multiple testing across 22 traits, two SNPs (Chr 22: 43,111,579 and Chr36: 8,058,628) were identified as significant in the admixture mapping (Fig. S10c–h). No variant was significantly associated with the 21 other traits used to define the morphs (Fig. S12).
Discussion
Using demographic models, we refuted the hypothesis of rapid sympatric differentiation of whitefish in Lake Tsahkal. Demographic analyses support an allopatric origin of whitefish morphs clearly predating the transplantation to the lake 50 years before sampling. This is not entirely surprising. While in fishes there are examples of rapid adaptive responses to novel environmental conditions on similar timescales, in most cases, these responses are subtle or have taken place over much longer time, commonly over hundreds of thousands of years (e.g., Albertson et al. 1999; Danley and Kocher 2001; Bolnick and Fitzpatrick 2007; Hudson et al. 2007, 2011 ).
Our results suggest that following a long phase of strict isolation, gene flow between divergent whitefish morphs was re-established during the last glacial retreat. Similar cases of secondary contact in post-glacial lakes have been reported from other northern temperate zone whitefish and they often led to hybridization (Bernatchez et al. 2010; Hudson et al. 2011; Rougeux et al. 2017, 2019b). The observed gradient of admixture suggests that the introduction of whitefish to the Lake Tsahkal created an artificial hybrid zone facilitating gene flow between littoral and pelagic morphs. The absence of genetic differentiation between the pelagic and profundal morphs was somewhat surprising. It is possible that these phenotypic differences are driven by plasticity but given our relatively low coverage of the whitefish genome it is also possible that we have missed important genetic differences between the two morphs underlying an adaptation to the different environments.
This study identifies five SNPs potentially associated with gonad weight variation. Gonad weight is an interesting trait because of its association with spawning time and thereby also with reproductive isolation among whitefish morphs. It is known that whitefish morphs have different spawning times, habitats, and size at maturity (Svärdson 1979; Vonlanthen et al. 2009; Bitz-Thorsen et al. 2020). The fact that the relative size of the gonads was largest in the littoral morph and smallest in the profundal morph (Fig. S5) makes sense as the former is an earlier breeder than the latter (Kahilainen et al. 2014). That differences in relative size of the gonads among the morphs were further mirrored in corresponding differences in frequency of five SNPs potentially associated with gonad size suggests that these SNPs are likely associated with breeding time differences among the morphs. Further weight on this inference is provided by the fact that in the lake whitefish (Coregonus clupeaformis), gonadosomatic index maps to chromosome 28 similarly to one of the SNPs identified by us (Gagnaire et al. 2013a). In closely related salmonids, chromosome 28 is related to sex determination in brown trout (Salmo trutta), body size in Arctic charr (Salvelinus alpinus), and links in multigene cluster with chromosome 22 in Atlantic salmon (Salmo salar) (Gharbi et al. 2005; Li et al. 2011; Norman et al. 2011). Furthermore, two of the SNPs associated with gonad weight variation displayed strong long-range LD (r2 > 0.9) across chromosomes (Chr. 22 and 36), a pattern that could be indicative of strong selection (Lewontin and Kojima 1960; Nei and Li 1973). Interchromosomal LD among loci under strong selection is not exceptional, as exemplified by strong patterns of interchromosomal LD associated with freshwater adaptation in sticklebacks (Fang et al. 2020). However, the possibility that the reported high long-range LD is due to genome misassemblies cannot be excluded. Selection for different spawning times may generate strong reproductive barriers and maintain reproductive isolation in syntopy as suggested by simulation models (Thibert-Plante et al. 2020) and empirical studies on European and Baltic flounders (Momigliano et al. 2017, 2018) and Atlantic cod (Fevolden and Pogson 1997; Hemmer-Hansen et al. 2013; Berg et al. 2016).
It is noteworthy that we found no SNP associated with gill raker number variation, a highly heritable trait playing a central role in foraging ecology and adaptive radiations of coregonids (Bernatchez 2004; Rogers and Bernatchez 2007; Kahilainen et al. 2011) nor any of the other measured traits. This is possibly a result of the small sample size of 41 admixed individuals and limited number of markers, which lowers the statistical power of GWA analyses (Hong and Park 2012). This could be particularly important if variation in gill raker number is a polygenic trait, which is notoriously difficult to map using GWA (Lin et al. 2021).
In conclusion, the results provide strong evidence that the divergence of the littoral and pelagic morphs in Lake Tsahkal predates their introduction to the lake. Although reproductive isolation among the two morphs is incomplete, their genetic differentiation suggests that spawning time differences have likely evolved in allopatry, and the morphs are currently maintaining reproductive isolation in the absence of other clear barriers to gene flow. The identification of five SNPs likely associated with gonad weight variation, a proxy for spawning time, lends support to this hypothesis and provides a starting point to identify causal loci underlying reproductive isolation. These analyses provide a starting point for further investigations on the genetic architecture of spawning time and the role of this trait in maintaining reproductive isolation, which should focus on expanding sample size, the number of studied populations, and the number of markers used.
ACKNOWLEDGMENTS
We thank T. Holopainen and P. Nieminen for their help in field work, M. Issakainen for help with laboratory work, and P. Kemppainen for advice and technical support in admixture mapping. The research is supported by the Academy of Finland (grants #129662, #134728, and #218343 to JM; #316294 to PM; #1140903 to KK), European Regional Development Fund (#A30205 to KK), the Finnish Cultural Foundation (Säätiöiden post doc-pooli; #00211290 to BF), and the Spanish Agencia Estatal de Investigación and European Union “NextGenerationEU/PRTR” (IJC2020-042611-I/MCIN/AEI/10.13039/501100011033 to PM).
CONFLICT OF INTEREST
The authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
JM and PM conceived the study. BF and PM conducted analyses. BF, JM, and PM wrote the manuscript. KK found phenotypic divergence, collected samples, measured ecomorphological data, and provided constructive comments on the manuscript. BF performed visualization. All authors approved the final version of the manuscript.
DATA ARCHIVING
Information about samples used is provided in Table S1. Raw sequence data have been uploaded to NCBI (PRJNA761129). The bioinformatic scripts used are deposited at Zenodo (https://doi.org/10.5281/zenodo.5519633).
REFERENCES
Associate Editor: J. Sachs
Handling Editor: Dr. T. Chapman




