Clinical and molecular characterization of patients with YWHAG-related epilepsy
Valentina Cetica and Tiziana Pisano contributed equally to this study.
YWHAG Study Group members are listed in the Appendix.
Abstract
Objective
YWHAG variant alleles have been associated with a rare disease trait whose clinical synopsis includes an early onset epileptic encephalopathy with predominantly myoclonic seizures, developmental delay/intellectual disability, and facial dysmorphisms. Through description of a large cohort, which doubles the number of reported patients, we further delineate the spectrum of YWHAG-related epilepsy.
Methods
We included in this study 24 patients, 21 new and three previously described, with pathogenic/likely pathogenic variants in YWHAG. We extended the analysis of clinical, electroencephalographic, brain magnetic resonance imaging, and molecular genetic information to 24 previously published patients.
Results
The phenotypic spectrum of YWHAG-related disorders ranges from mild developmental delay to developmental and epileptic encephalopathy (DEE). Epilepsy onset is in the first 2 years of life. Seizure freedom can be achieved in half of the patients (13/24, 54%). Intellectual disability (23/24, 96%), behavioral disorders (18/24, 75%), neurological signs (13/24, 54%), and dysmorphisms (6/24, 25%) are common. A genotype–phenotype correlation emerged, as DEE is more represented in patients with missense variants located in the ligand-binding domain than in those with truncating or missense variants in other domains (90% vs. 19%, p < .001).
Significance
This study suggests that pathogenic YWHAG variants cause a wide range of clinical presentations with variable severity, ranging from mild developmental delay to DEE. In this allelic series, a genotype–phenotype correlation begins to emerge, potentially providing prognostic information for clinical management and genetic counseling.
Key points
- The phenotypic spectrum of YWHAG-related disorders ranges from mild developmental delay to developmental and epileptic encephalopathy.
- Language impairment is common; comorbidities include ataxia, tremors, and behavioral disorders, mainly autistic features, ADHD, and aggressive behavior.
- Seizure freedom can be achieved in most patients.
- A preliminary genotype–phenotype correlation begins to emerge, as the most severe phenotypes cluster in patients with variants in the ligand-binding domain.
1 INTRODUCTION
The YWHAG gene (tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein gamma, Mendelian Inheritance in Man [MIM] 605356) encodes a member of the highly conserved 14-3-3 protein family, which is involved in multiple intracellular signaling pathways. Through binding to their targets, 14-3-3 proteins contribute to the regulation of a large spectrum of biological processes including signal transduction, cell cycle, transcription, and apoptosis.1
Given their expression in the developing brain, especially the cortex, and their crucial role in neuronal development, 14-3-3 proteins are involved in several human diseases, mainly neurodevelopmental, neurodegenerative, and neuropsychiatric disorders.2-4
The isoform gamma, named YWHAG, is highly expressed in the brain, skeletal muscle, and heart. Animal models indicate that alterations in this gene have consequences on neurologic development and cardiac function. Knockdown of Ywhag1 in zebrafish results in reduced brain size and enlarged diameter of the heart tube with increased risk for cardiac arrhythmia.5 In utero ablation in Ywhag−/− mice, with consequent alteration of Ywhag levels, leads to delayed neuronal migration in the developing brain.6
Komoike et al.5 studied Ywhag in zebrafish and suggested that haploinsufficiency for the human orthologue YWHAG, which maps in the 7q11.23 interval that is deleted in Williams–Beuren syndrome (MIM 194050), was related to infantile seizures and cardiomyopathy in these patients. Guella et al.7 described heterozygous YWHAG single nucleotide variants in seven patients with early onset epilepsy. Overall, 13 missense, one nonsense, and one frameshift variant have been reported in patients with epilepsy or developmental and epileptic encephalopathy (DEE).
We characterized clinical manifestations, electroclinical features, epilepsy course, and outcome of 24 novel patients with pathogenic or likely pathogenic YWHAG variant alleles. Based on observations of the newly reported patients described herein and those previously reported, we expand the overall spectrum of phenotypes associated with YWHAG variants.
2 MATERIALS AND METHODS
We retrospectively collected clinical and molecular data from 24 patients with pathogenic/likely pathogenic YWHAG variants. Patients were diagnosed at Meyer Children's Hospital Istituto di Ricovero e Cura a Carattere Scientifico (IRCCS) (n = 2) or recruited through GeneMatcher (n = 8),8 collaborations with national and international epilepsy centers (n = 8), the Epi25 consortium (n = 5),9 and Decipher (n = 1).10 Three patients had been previously published (Patients 6, 10, and 11)11-13 and are rephenotyped in this study. Referring physicians completed a questionnaire with comprehensive genetic and clinical information, including electroencephalographic (EEG) and neuroimaging data. Because patients were followed at different centers and had variable ages at the time of study, cognitive level was evaluated using different methods, including the Griffiths Mental Development Scale,14 the Wechsler Intelligence Scale for Children,15 and the Wechsler Adult Intelligence Scale–Revised,16 as well as adaptive behavioral criteria. Autism was diagnosed using Diagnostic and Statistical Manual of Mental Disorders, 5th edition.17
2.1 Literature review
For literature review of previously published patients carrying pathogenic or likely pathogenic YWHAG variants, we searched PubMed for all publications including YWHAG variants, using as MeSH terms “YWHAG” and either “mutations” or “variants”, from the date of the first published patient5 until May 2023. We excluded patients with genomic deletions including other genes besides YWHAG. The reference lists of the selected publications were examined for additional potentially relevant publications. Genetic inclusion criteria were missense or truncating variants in YWHAG, classified as pathogenic or likely pathogenic according to the American College of Medical Genetics and Genomics guidelines.18 The identified variants were cross-checked with YWHAG variants reported in the Human Gene Mutation Database, and data on the associated neurological phenotypes were extracted when available. We eventually reviewed 24 previously reported patients harboring 15 different pathogenic or likely pathogenic YWHAG variants.
2.2 Standard protocol approvals, registrations, and patient consents
This study was approved by the Pediatric Ethics Committee of the Tuscany Region, and informed consent was obtained from patients, parents, or guardians.
2.3 Genetic study
We studied 24 patients with de novo (n = 19), inherited (n = 3), or unknown inheritance (n = 2) pathogenic/likely pathogenic variants in YWHAG. Molecular genetic testing was performed using exome (Patients 1–7 and 10–24) or multigene panel sequencing (Patients 8 and 9). Patients were identified across 20 institutions with data shared through the Epi25 consortium and match-making strategies (GeneMatcher, international collaborations, Decipher). Detailed information about sequencing methods is provided in the Supplementary Methods. Five more patients (three via international collaborations and one each via the Epi25 consortium and GeneMatcher) had variants in YWHAG about whose pathogenicity we had low confidence, hence we analyzed their data separately.
For six patients (Patients 10–12, 18, 19, 21), exome sequencing (ES) was performed as part of the Epi25 project. These samples were sequenced as singletons at the Broad Institute of Harvard and the Massachusetts Institute of Technology.
For two patients (Patients 4 and 13), genomic DNA was obtained from peripheral blood from the proband and parents, and ES was performed at the Baylor College of Medicine Human Genome Sequencing Center, as previously described.19, 20 For the 16 remaining patients, sequencing was performed at different centers within research or diagnostic settings. All variants were tested with six bioinformatic tools (SIFT, Polyphen2 HumVar, MutationTaster, MutationAssessor, FATHMM, FATHMM MKL) included in the VarSeq software (Golden Helix).
2.4 Statistical analysis
We applied the two-sided Fisher exact test, with statistical significance set to p < .05, to explore possible associations between variant location or type and clinical features (primary syndromic diagnosis, response to treatment, developmental delay and intellectual disability, language impairment, neurological findings, behavioral abnormalities, and dysmorphic features) in the whole cohort of new and previously described patients.
3 RESULTS
3.1 Genetic analysis
Genetic and clinical information on 24 individuals (21 new; three previously published) harboring pathogenic/likely pathogenic YWHAG variants are summarized in Table 1 and presented in more detail in Table S1. These 24 patients harbored 12 different YWHAG variants, which occurred de novo in 19 of them, were inherited from an unaffected mother in three (Patients 1, 8, 9), and remained of undetermined origin in two, whose parents were unavailable for segregation analysis (Patients 5, 11). Among the 12 pathogenic/likely pathogenic variants, nine were missense (9/12, 75%), two frameshift (2/12, 17%), and one nonsense (1/12, 8%). The three patients with a variant inherited from an unaffected parent carried a frameshift variant. Arginine 57 and arginine 132 represent two hotspot amino acid residues affected by multiple missense changes (p.Arg57Cys, p.Arg57Gly, p.Arg57His, p.Arg132His, p.Arg132Cys, and p.Arg132Gly), recurring in six and in 11 patients, respectively. Except for the p.Arg132Gly, all these changes had been previously described. One patient (Patient 10, also included in Epi4K 2013) carried the c.387C>G p.(Asp129Glu) missense substitution and another patient (Patient 23) the c.619G>A p.(Glu207Lys), a variant previously found in a different individual (Patient 48).21
| Genetic and clinical feature | New cohort: Patients/patients with available data, n (%) | Literature: Patients/patients with available data, n (%) | Total: Patients/patients with available data, n (%) |
|---|---|---|---|
| Variant type | |||
| Missense | 20/24 (83) | 17/24 (71) | 37/48 (77) |
| Frameshift | 3/24 (13) | 6/24 (25) | 7/488 (15) |
| Nonsense | 1/24 (4) | 1/24 (4) | 4/48 (8) |
| De novo variant | 19/22 (79) | 16/21 (67) | 35/43 (81) |
| Sex | |||
| M | 14/24 (58) | 12/22 (55) | 24/46 (52) |
| F | 10/24 (42) | 10/22 (45) | 22/46 (48) |
| Age, median (range) | 17 years (3 years 6 months–67 years) | 13 years 11 months (3 years–40 months) | 15 years (3 years–67 years) |
| Primary diagnosis | |||
| DEE | 20/24 (83) | 2/15 (13) | 22/39 (56) |
| GE | 4/24 (17) | 12/15 (80) | 16/39 (41) |
| Focal epilepsy | 1/15 (7) | 1/39 (3) | |
| Seizure onset, median (range) | 10 months (1 months–2 years) | 2 years (2 months–16 years) | 1 years 5 months (1 month–16 years) |
| Predominant seizure type at follow-up | Myoclonic: 15/23 (65) | Myoclonic: 3/9 (33) | Myoclonic: 18/32 (56) |
| Seizure triggers | 14/23 (58) | 9/9 (100) | 22/32 (69) |
| Fever | 11/14 (79) | 8/9 (89) | 19/22 (86) |
| Abnormal EEG at onset | 25/21 (71) | 3/3 (100) | 18/24 (75) |
| Abnormal EEG at last evaluation | 18/22 (82) | 5/10 (50) | 24/32 (75) |
| ASM | |||
| Polytherapy | 20/24 (83) | 7/19 (37) | 27/43 (63) |
| VPA monotherapy | 4/24 (17) | 8/19 (42) | 12/43 (28) |
| No therapy | 4/19 (21) | 4/43 (9) | |
| Response to treatment | SF: 13/24 (54) | SF: 15/19 (79) | SF: 28/43 (65) |
| DD/ID | 23/24 (96) | 13/21 (62) | 36/45 (80) |
| Mild/moderate | 13/23 (57) | 13/13(100) | 26/36 (72) |
| Severe | 10/23 (43) | 0 | 10/36 (28) |
| Language impairment | 21/24 (88) | 7/12 (58) | 30/36 (86) |
| Neurological findings | 13/23 (54) | 5/5 (100) | 18/28 (64) |
| Myoclonic jerks during sleep, nonepileptic | 11/22 (50) | 1/1 (100) | 12/23 (52) |
| Sleep disorders | 9/23 (39) | NA | 9/23 (39) |
| Behavioral abnormalities/ASD | 16/24 (67) | 4/6 (67) | 20/30 (67) |
| Abnormal brain MRI | 4/24 (17) | 4/15 (27) | 8/39 (21) |
| Dysmorphic features | 6/24 (25) | 9/13 (69) | 15/37 (41) |
- Abbreviations: ASD, autism spectrum disorder; ASM, antiseizure medication; DD/ID, developmental delay/intellectual disability; DEE, developmental and epileptic encephalopathy–myoclonic atonic epilepsy; EEG, electroencephalogram; F, female; GE, generalized epilepsy; M, male; MRI, magnetic resonance imaging; NA, not available; SF, seizure-free; VPA, valproic acid.
In the five remaining patients, we identified four novel variants (c.578C>T p.[Ala193Val], c.698G>A p.[Trp233*], c.89dup p.[Thr31Aspfs*5], and c.187_188insC p.[Ile63Thrfs*3]).
None of the variants was present in the gnomAD database (v2.1.1, accessed May 2023), and all occurred at highly conserved amino acid residues. Arginine 57 and 132 were located inside a hydrophobic groove involved in ligand binding. p.Asp129Glu was predicted to be involved in interactions with ligands through a new hydrogen bond with phosphopeptides.22 Alanine 193 and glutamate 207 mapped in a protein region that could be involved in the interaction between two α-helical structures, thus contributing to stabilizing the three-dimensional conformation.
The p.(Thr31Aspfs*5), p.(Ile63Thrfs*3), and p.(Trp233*) frameshift variants introduce a premature stop codon, leading to early protein synthesis termination. The role of YWHAG truncating variants is controversial. YWHAG is classified as a loss of function (LoF) intolerant gene (probability of being LoF intolerant = .96), but it is also predicted to escape nonsense-mediated decay (NMD− gene),23 and it is unclear whether these truncating variants act via an LoF effect. All known YWHAG variants, including those identified in this and previous reports, are depicted in Figure 1.

3.2 Phenotypic characterization
Clinical information of the patients included in our cohort is summarized in Table 1 and detailed in Table S1. Fourteen patients were males and 10 females (gender ratio male/female = 1.4/1), with an age at last follow-up ranging from 3 years 6 months to 67 years (mean = 17 year 6 months). All patients had epilepsy, with a median age at seizure onset of 10 months (range = 1 month to 2 years). The main diagnostic category was DEE, observed in 20 patients (20/24, 83%); the remaining four had generalized epilepsy (4/24, 17%). At onset, generalized tonic–clonic seizures were observed in 14 patients (14/23, 61%), absence seizures in three (3/23, 13%), myoclonic seizures in two (2/23, 9%), infantile spasms in one (1/23, 4%), and atypical absences in one (1/23, 4%). Two patients (2/23, 9%) had manifested multiple seizure types since epilepsy onset. During follow-up, 15 patients had presented myoclonic seizures (15/23, 65%), associated with other types of seizures in 12 of them and as the only type of seizure in the remaining three.
In eleven patients (11/23, 48%), seizures were triggered by fever. Other reported triggers were photic (1/23, 4%) and tactile and sound stimuli (1/23, 4%). One patient (1/23, 4%) had catamenial epilepsy.
We obtained data on EEG at onset for 21 patients (21/24, 87%). In 15 patients (15/21, 71%), EEG showed generalized spike and waves or both focal and generalized abnormalities; normal EEG recordings were reported at onset in six patients (6/21, 29%). EEG at last evaluation, available for 22 of 24 (92%) patients, exhibited slow background activity in eight (8/22, 36%) and a combination of focal, multifocal, or generalized spike wave and/or polyspike discharges (16/22, 73%) that increased during sleep in five of 22 patients (23%). In four patients (4/22, 18%), EEG was normal. All patients were treated with antiseizure medications (ASMs), consisting of valproic acid (VPA) monotherapy (4/24, 17%) or polytherapy including combinations of different drugs (20/24, 83%). Thirteen patients (13/24, 54%) are currently seizure-free: six after ASM withdrawal (6/13, 46%), four on VPA monotherapy (4/13, 31%), and three on a combination therapy (3/13, 23%). In nine of the 20 patients with DEE, language impairment and intellectual disability persisted while epilepsy went into remission.
Twenty-three patients (23/24, 96%) exhibited intellectual disability, ranging from mild (6/23, 26%) to moderate (7/23, 30%) and severe (10/23, 4%). Delayed milestones were a frequent presentation in our cohort, with nine patients (9/23, 39%) showing delayed walking and 21 (21/24, 88%) language impairment, from mild (7/21, 33%) to moderate (5/21, 24%) and severe (9/21, 43%). Only three patients developed normal language skills (3/24, 12%). Other neurological features, described in 13 patients (13/24, 54%), included tremor (8/13, 62%), ataxia (8/13, 62%), and clumsiness (7/13, 54%). Eight patients (8/13, 62%) manifested more than one neurological sign. Nonepileptic myoclonic jerks during sleep were reported in 11 patients (11/22, 50%) and sleep disorders (fragmented sleep, difficulty in settling at night, and nocturnal awakenings) in nine (9/23, 39%). Autism spectrum disorder was diagnosed in 13 of 24 (54%) patients, attention-deficit/hyperactivity disorder (ADHD) in eight of 24 (33%), and aggressive behavior in three of 24 (12%). Brain magnetic resonance imaging (MRI) was available for all patients and was reported to be normal in 20 (20/24, 83%) and abnormal in four (4/24, 17%). MRI alterations were variable and included mildly dilated frontal horns, cortical atrophy, arachnoid cyst, and Arnold–Chiari I malformation. Facial dysmorphisms were reported in six patients (6/24, 25%).
3.3 Literature review
Thirteen different YWHAG variants had been previously described in 24 patients. Detailed information of these patients is presented in Table 1 and Table S2.
3.4 Analysis of all patients
Combining our findings with previous reports, 48 patients with a pathogenic or likely pathogenic variant in YWHAG can now be identified. Genetic, clinical, EEG, and MRI data of all the 48 patients are summarized in Table 1 and Figure 2.
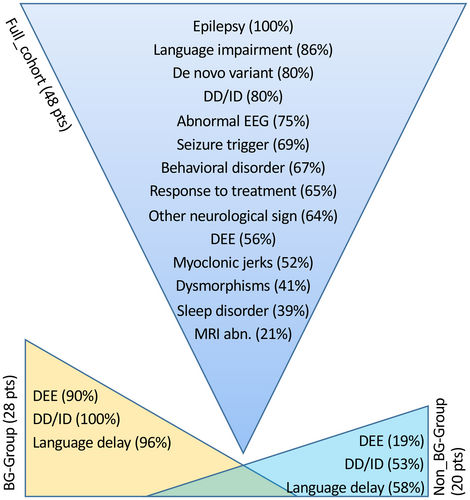
3.5 Genotype–phenotype correlations
A possible genotype–phenotype correlation emerged when dividing the full set of 48 patients into two groups, based on the location of their variants. The first group, the BG-group, corresponded to recurrent variants located in the binding groove (BG) domain (28 patients). The second group, the non-BG-group, harbored variants located in other domains of the protein (20 patients). Two-sided Fisher exact test (significance set at p < .05) indicates that a higher proportion of patients in the BG-group had a DEE (90% in BG-group vs. 19% in non-BG-group, p < .001), with developmental delay—intellectual disability (100% in BG-group vs. 53% in non-BG-group, p < .001) and language impairment (96% BG-group vs. 58% non-BG-group, p = .006; Figure 2). A trend toward a higher proportion of neurological comorbidities in the BG-group is also apparent, although this association does not reach the confidence threshold (76% in BG-group vs. 29% in non-BG-group, p = .06). We then compared patients with missense variants to those carrying truncating (nonsense and frameshift) variants and found that developmental delay and intellectual disability (94% missense vs. 36% truncating, p < .001), language impairment (93% missense vs. 20% truncating, p < .001), and neurological comorbidities (72% missense vs. 0 truncating; p = .03) were more common in patients carrying missense variants. A higher percentage of patients with truncating variants became seizure-free (56% missense vs. 91% truncating, p = .065), although these data need to be confirmed in a wider cohort. All patients with DEE carried a missense variant.
3.6 Patients with variants of uncertain significance
Five novel patients harbored four different missense YWHAG variants that did not fully satisfy the criteria for classification as pathogenic or likely pathogenic (Table S3), hence we analyzed them separately. These were four unrelated patients plus the symptomatic father of one of them. The c.176C>T p.(Ser59Phe) and the c.517T>G p.(Leu173Val) variants are absent from the GnomAD population database, whereas the c.87C>G p.(Asn29Lys) and the c.431C>T p.(Ala144Val) variants occur in one of 226 866 and four of 251 470 alleles, respectively. The bioinformatic tools we applied were not concordant in defining the pathogenic effect of these variants. We could demonstrate that all variants were inherited from a mildly affected parent, except p.(Leu173Val; Patient 53), where segregation analysis could not be performed. None of the patients was diagnosed with DEE. The diagnostic categories were generalized epilepsy (Patients 49, 51, 52) and global developmental delay without seizures (Patient 50). Two patients with epilepsy were seizure-free, Patient 49 for 2 years on VPA monotherapy and Patient 52 for 34 years without ASM. Language was reported to be normal in these patients. Autism spectrum disorder was observed in one patient (Patient 50).
4 DISCUSSION
The initial reports of individuals with YWHAG variants described an early onset epileptic encephalopathy with prevalently myoclonic seizures, intellectual disability, developmental delay, and facial dysmorphisms.7, 11 More recent reports included milder phenotypes of myoclonic epilepsy and febrile seizures.22, 24 By adding a consistent number of new patients, we expand the characterization of phenotypes associated with YWAHG pathogenic/likely pathogenic variants and illustrate emerging genotype–phenotype correlations for this gene.
Our data substantiate specific features observed in previously described patients and highlight additional aspects. Our series confirms a YWHAG-associated phenotype encompassing a wide spectrum of neurodevelopmental disorders, from mild developmental delay and epilepsy to severe DEE (Figure 2), and contributes novel insights about EEG features, clinical epilepsy, and its outcome. As previously reported, epilepsy is the most frequent clinical presentation, with onset in the first 2 years of life. Seizures are mainly generalized tonic–clonic, triggered by fever in 46% of our patients. Over time, myoclonic seizures become prominent (65%). EEG data in our cohort show, in most patients, slow background activity and a combination of focal and multifocal abnormalities and generalized polyspikes or spike and wave discharges. Seizure control is achieved in 54% of patients, with VPA monotherapy (23%) or a combination of drugs (77%), even when patients present as DEE at onset. These observations reinforce previous findings wherein pharmacological control of seizures was reported in several patients.7, 21, 22
Developmental delay and intellectual disability of variable severity were present in most of our patients (96%), but were reported in only 60% of previously described series. This discrepancy may arise from the inclusion in the literature dataset of several patients carrying truncating variants and exhibiting mild manifestations, most of whom belonged to a single large family.22 Additional observations including a more mixed population are needed to clarify the spectrum of cognitive impairment associated with YWHAG variants. Language impairment is a common feature. Comorbidities, not systematically explored in previous reports, include behavioral disorders (67%), mainly autistic features, ADHD, and aggressive behavior. Nonepileptic myoclonic jerks during sleep were reported in 50% of our patients and sleep disorders (fragmented sleep, difficulty in settling at night, and nocturnal awakenings) in 39%. Fifty-four percent of patients exhibited neurological signs such as ataxia, tremors, clumsiness, and hypotonia. Variable brain MRI alterations, not suggesting a recurrent pattern of abnormality, were identified in 17% of our patients (4/24, 17%) and in 27% (4/15, 27%) of those previously reported, for whom, however, this information was not available in 38%. Lack of information does not allow discerning whether MRI was not performed or unrevealing, leaving open the possibility that a systematic analysis in a large cohort, ideally by applying morphometric methods, might reveal a common pattern of abnormality, despite nonspecific qualitative findings.25, 26
Dysmorphisms, described in 69% of previously published patients, were observed in only 25% of those included in this report, and no recognizable recurrent pattern could be identified in this and previous studies. However, mild dysmorphisms in a rare condition are easily underreported, as they may escape recognition if patients are not seen by expert clinical geneticists and are reported via an international collaboration in which each center contributes single or very few observations, making interindividual comparison impossible.
Twelve distinct YWHAG variants are present in our series, totaling 19 when including those reported previously (Tables S1 and S2), most being missense variants. Two mutational hotspots exist: Arg132 and Arg57. These highly conserved amino acids, along with Tyr133, previously identified in two unrelated patients, form a positively charged pocket within a hydrophobic groove (BG), which is functionally important for the binding of the 14-3-3γ protein with different ligands. 14-3-3γ, encoded by YWHAG, like the other 14-3-3 proteins, functions almost exclusively in its dimeric form and exerts its function through binding with phosphopetides.27 In the group of newly and previously described patients with variants affecting one of the three hotspot variants, the BG-group, the prevalence of DEEs is statistically significant (90% in BG-group vs. 19% in non-BG-group, p < .001), and associates with developmental delay and intellectual disability (100% in BG-group vs. 53% in non-BG-group, p < .001) and language impairment (96% in BG-group vs. 58% in non-BG-group, p = .006). These findings suggest that patients carrying an alteration in the key domain responsible for ligand binding are more severely affected. It is likely that the three hotspot alterations have a dominant negative effect that impairs phosphopetides binding and disrupts the regulatory role of 14-3-3γ during neuronal development, thus resulting in more severe neurological impairment. Conversely, an alteration at a different site could destabilize the dimeric structure without impeding its full formation and function. All variants falling into the BG-group are de novo and missense.
When comparing patients' phenotypes associated with missense (37 patients) versus truncating variants (11 patients), it becomes obvious that none of the patients with truncating variants received a diagnosis of DEE. In addition, the proportion of those with developmental delay/intellectual disability (94% of missense vs. 36% of truncating, p < .001), language impairment (93% of missense vs. 20% of truncating, p < .001), and neurological comorbidities (71% of missense vs. 0 truncating, p = .03) is less represented in the group with truncating variants. This milder phenotype could be due to haploinsufficiency, which leaves intact the functionality of the residual encoded protein, as opposed to the putative dominant negative effect exerted by missense variants. These data need to be confirmed in a wider series and corroborated by functional studies probing the effect of these variants on protein quantity and function.
Although not statistically significant, data on treatment response suggest that the percentage of patients who respond to ASMs seems to correlate with the type and location of genetic variants, and good seizure control is often achieved if variants fall outside the hydrophobic BG (p = .09) or are truncating (p = .02). This observation, if confirmed, might help future precision medicine approaches.
The five YWHAG variants classified as variants of uncertain significance (VUS) were associated with heterogeneous epilepsy phenotypes in four, including absence seizures (Patient 49), generalized tonic–clonic seizures triggered by fever (Patients 51, 52), and developmental delay without epilepsy (Patient 50). Except for Patient 50, who has global developmental delay and facial dysmorphisms, the remaining four patients exhibited normal development and language skills. These phenotypic characteristics seem to overlap with the milder side of the spectrum of YWHAG-related disease. Further observations in a wider cohort of patients with VUS might support the hypothesis that hypomorphic variants exist in this gene.
5 CONCLUSIONS
This cohort highlights the wide range of clinical presentations and severity in YWHAG-related disorders, which vary from mild developmental delay and epilepsy to DEE. Epilepsy onset is within the first 2 years, developmental delay/intellectual disability occurs in most patients, and language impairment is common. Other comorbidities, such as behavioral abnormalities and dysmorphic features, are variably manifested, making this syndrome difficult to recognize on clinical grounds only. A genotype–phenotype correlation begins to emerge, which reflects distinct molecular mechanisms and may prove helpful for clinical management, prognostication, genetic counseling, and precision medicine.
AUTHOR CONTRIBUTIONS
Study concept and design: Valentina Cetica, Tiziana Pisano, and Renzo Guerrini. Acquisition, analysis, and interpretation of data: Valentina Cetica, Tiziana Pisano, Renzo Guerrini, Gaetan Lesca, Dana Marafi, Laura Licchetta, Florence Riccardi, Davide Mei, Hon-yin B. Chung, Allan Bayat, Meena Balasubramanian, Daniel H. Lowenstein, Milda Endzinienė, Maha Alotaibi, Nathalie Villeneuve, Julia Jacobs, Bertrand Isidor, Roberta Solazzi, Nicolette S. den Hollander, Dragan Marjanovic, Christelle Rougeot-Jung, Julien Jung, Marion Lesieur-Sebellin, Andrea Accogli, Vincenzo Salpietro, Nebal W. Saadi, Eleni Panagiotakaki, Thomas Foiadelli, Sylvia Redon, Meng-Han Tsai, Francesca Bisulli, Trine B. Hammer, James R. Lupski, and Elena Parrini. All authors critically reviewed the manuscript and approved the final version for publication.
ACKNOWLEDGMENTS
The authors thank all patients and families. Meyer Children's Hospital IRCCS, IRCCS Istituto delle Scienze Neurologiche di Bologna, Fondazione IRCCS Istituto Neurologico Carlo Besta, University Hospital of Lyon, Hospital of Lithuanian, and University of Health Sciences Kauno Klinikos are full members of the European Reference Network (ERN) EpiCARE. Meyer Children's Hospital IRCCS is a full member of the ERN ITHACA. F.R. is a member of the expertise center of the French national network AnDDI-Rares. N.V. and M. Milh are members of the expertise center on rare epilepsies of the French national network DefiScience. We thank the Epi25 principal investigators, local staff from individual cohorts, and all of the patients with epilepsy who participated in the study for making possible this global collaboration and resource to advance epilepsy genetics research. This work is part of the Centers for Common Disease Genomics (CCDG) program, funded by the National Human Genome Research Institute (NHGRI) and the National Heart, Lung, and Blood Institute. CCDG-funded Epi25 research activities at the Broad Institute, including genomic data generation in the Broad Genomics Platform, are supported by NHGRI grant UM1 HG008895 (principal investigators: Eric Lander, Stacey Gabriel, Mark Daly, Sekar Kathiresan). The Genome Sequencing Program efforts were also supported by NHGRI grant 5U01HG009088–02. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We thank the Stanley Center for Psychiatric Research at the Broad Institute for supporting the genomic data generation efforts.
CONFLICT OF INTEREST STATEMENT
J.R.L. has stock ownership in 23andMe, is a paid consultant for Genome International, and is a coinventor on multiple US and European patents related to molecular diagnostics for inherited neuropathies, eye diseases, genomic disorders, and bacterial genomic fingerprinting. The Department of Molecular and Human Genetics at Baylor College of Medicine receives revenue from clinical genetic testing conducted at Baylor Genetics Laboratories. The other authors have no potential conflicts to disclose.
FUNDING INFORMATION
R.G. is supported by Tuscany Region Call for Health 2018 (grant DECODE-EE) and the Optical Brain Mapping project by Fondazione Cassa di Risparmio di Firenze. Meyer Children's Hospital IRCCS is funded by Current Research Funding (Italian Ministry of Health). M.B. is funded by Medical Research Council (MR/V037307/1) academic salary support. The Genome Sequencing Program efforts for Epi25 were supported by National Human Genome Research Institute grant 5U01HG009088. A supplemental grant for Epi25 phenotyping was supported by Epi25 Clinical Phenotyping R03, National Institutes of Health (R03NS108145). This study was supported in part by the US National Human Genome Research Institute (NHGRI) and National Heart, Lung, and Blood Institute to the Baylor-Hopkins Center for Mendelian Genomics (UM1 HG006542) and Baylor College of Medicine Genomic Research to Elucidate the Genetics of Rare Diseases (U01 HG011758), US National Institute of Neurological Disorders and Stroke (R35NS105078 to J.R.L.), and Muscular Dystrophy Association (512 848 to J.R.L.). Da.M. was supported by a Medical Genetics Research Fellowship Program through the US National Institutes of Health (T32 GM007526-42).
ETHICS STATEMENT
This study was approved by the Pediatric Ethics Committee of the Tuscany Region, and informed consent was obtained by patients, parents, or legal guardians.
APPENDIX: YWHAG STUDY GROUP
Lena Alotaibi, MD,1 Irene Ambrosetti, MD,2 Séverine A. Bellanger, MD, PhD,3,4 Barbara Castellotti, BS,5 Mara Cavallin, MD, PhD,6 Joshua C. K. Chan, MBBS,7 Nicolas Chatron, MD, PhD,8,9 Julie Chavany, MD,10 Benjamin Cogne, PharmD, PhD,11,12 Jasmine L. F. Fung, BBiomedSc MD,7 Cathrine E. Gjerulfsen, MD,13 Tiziana Granata, MD,5 Anne Guimier, MD, PhD,14 Isabella Herman, MD, PhD,15 Chen-Jui Ho, MD,16 Claudia Mandorlini, MS,6 Mathieu Milh, MD, PhD,17,18 Raffaella Minardi, PhD,19 Francesca Montanari, MD,20 Jill A. Rosenfeld, MS,15,21 Rikke S. Moller, PhD,22 Francesca F. Operto, MD,23 Jennifer E. Posey, MD, PhD,15 Claudia A. L. Ruivenkamp, PhD,24 Elise Sacaze, MD, PhD,3,4 Viola Santi, MD,25 Salvatore Savasta, MD,26 Renaud Touraine, MD, PhD,27 Birute Tumiene, MD, PhD,28 Kevin Uguen, MD, PhD,3,4,29 Laurent Villard, MD18
1College of Medicine, King Saud bin Abdul-Aziz University for Health Sciences, Riyadh, Saudi Arabia; 2Medical Genetics Unit, Azienda Ospedaliero–Universitaria di Bologna, Bologna, Italy; 3Service de Génétique Médicale, CHU de Brest, Brest, France; 4Centre de Référence Déficiences Intellectuelles de Causes Rares, Brest, France; 5Department of Diagnostics and Technology, Unit of Medical Genetics and Neurogenetics, Fondazione IRCCS Istituto Neurologico Carlo Besta, Milan, Italy; 6Neuroscience Department, Meyer Children's Hospital IRCCS, Florence, Italy; 7Department of Paediatrics and Adolescent Medicine, School of Clinical Medicine, LKS Faculty of Medicine, University of Hong Kong, Hong Kong, China; 8University of Lyon 1, CNRS, INSERM, Physiopathologie et Génétique du Neurone et du Muscle, UMR5261, U1315, Institut NeuroMyoGène, Lyon, France; 9Department of Genetics, University Hospitals of Lyon, HCL, Lyon, France; 10CHITS, Hôpital Ste Musse, Service de Pédiatrie, Toulon, France; 11Service de Génétique Médicale, CHU Nantes, Nantes, France; 12Université de Nantes, CNRS, INSERM, l'Institut du Thorax, CHU Nantes, Nantes, France; 13Department of Epilepsy Genetics and Personalized Medicine, Danish Epilepsy Center, Dianalund, Denmark; 14Department of Genomic Medicine of Rare Disorders, Necker Hospital, APHP Center, University Paris Cité, Paris, France; 15Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, Texas, USA; 16Department of Neurology, Kaohsiung Chang Gung Memorial Hospital, Kaohsiung, Taiwan; 17Department of Pediatric Neurology, AP-HM, La Timone Children's Hospital, Marseille, France; 18Faculté de Médecine Timone, Aix-Marseille University, INSERM, MMG, U1251, Marseille, France; 19IRCCS Istituto delle Scienze Neurologiche di Bologna, Bologna, Italy; 20Medical Genetics Unit, IRCCS Azienda Ospedaliero–Universitaria di Bologna, Bologna, Italy; 21Baylor Genetics Laboratory, Houston, Texas, USA; 22Department of Epilepsy Genetics and Personalized Medicine, Danish Epilepsy Center, University of Southern Denmark, Filadelfia, Denmark; 23Department of Science of Health, School of Medicine, University Magna Greacia of Catanzaro, Catanzaro, Italy; 24Department of Clinical Genetics, Leiden University Medical Center, Leiden, the Netherlands; 25Clinica Pediatrica, Fondazione IRCCS Policlinico San Matteo Pavia, Italy; 26Pediatric Clinic and Rare Diseases, Department of Medical Sciences and Public Health, University of Cagliari, Cagliari, Italy; 27Department of Medical Genetics, University Hospital of Saint-Etienne, Saint-Etienne, France; 28Vilnius University, Faculty of Medicine, Institute of Biomedical Sciences, Vilnius, Lithuania; 29University of Brest, Inserm, EFS, UMR 1078, GGB, Brest, France
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.




