Influence of the Gut Microbiota on Acute Ischemic Stroke Functional Outcomes at Three Months
Funding: This study has been funded by Instituto de Salud Carlos III (ISCIII) (grant numbers PI18/01338, PI20/00925), ERA-NET NEURON (grant number AC19/00106), RICORS-ICTUS: Red de Investigación Cooperativa Orientada a Resultados en Salud—Enfermedades Vasculares Cerebrales (grant number RD21/0006/0006). M. Lledós is funded by a PFIS Contract (Contratos Predoctorales de Formación en Investigación en Salud, grant number FI19/00309) from ISCIII. C. Gallego-Fabrega is supported by a Sara Borrell Contract (grant number CD20/00043) from ISCIII and the European Regional Development Fund (ISCIII-ERDF).
Miquel Lledós and Luís Prats-Sánchez contributed equally to this work.
ABSTRACT
Background
Functional recovery from ischemic stroke (IS), the main cause of adult disability worldwide, is influenced by many factors, and a portion of interindividual variability remains unexplained.
Methods
Observational study in a tertiary stroke centre of patients with IS analyzed using shotgun metagenomic sequencing (January 2020–March 2022). Functional outcomes were assessed according to modified Rankin Scale (mRS) scores 3-months post-IS, considering 0–2 favorable and 3–6 unfavorable. The causal relationship between several bacteria and post-IS outcomes was explored via two-sample Mendelian randomization (MR) analyses using Genome-Wide Association Analysis (GWAS) summary statistics.
Results
Comparing 128 patients with favorable and unfavorable post-IS functional outcomes, β-diversity analysis showed a separation in microbial structure, and α-diversity measures revealed greater bacterial richness in the favorable outcomes group. Taxonomic profiling of the samples showed that a greater abundance of pathogenic bacteria (e.g., Pseudomonas, Finegoldia, Porphyromonas) was associated with an unfavorable outcome. Functional profiling of the samples revealed differences in the ethylbenzene degradation pathway and in 16S rRNA (uracil1498-N3)-methyltransferase. MR confirmed increased pyruvate levels to be causally associated with post-IS favorable outcomes (β = −0.50, 95% CI: −0.91, −0.10).
Conclusions
Our study points to gut microbiota differences in patients with unfavorable versus favorable 3-month post-IS outcomes. Patients with unfavorable outcomes presented gut microbiota dysbiosis and alterations in multiple metabolic pathways.
Trial Registration: This study was registered on 3 October 2021 with https://clinicaltrials.gov. Access number: NCT04795687
Abbreviations
-
- ACE
-
- abundance-based coverage estimator
-
- BMI
-
- body mass index
-
- EC
-
- Enzyme Commission
-
- GISCOME
-
- Genetics of Ischemic Stroke Functional Outcome
-
- GSEA
-
- gene set enrichment analysis
-
- GWAS
-
- genome-wide association study
-
- IS
-
- ischemic stroke
-
- IV tPA
-
- intravenous tissue-type plasminogen activator
-
- IVW
-
- inverse-variance weighted
-
- KEGG
-
- Kyoto Encyclopedia of Genes and Genomes
-
- LDA
-
- linear discriminant analysis
-
- LEfSe
-
- LDA effect size
-
- MR
-
- Mendelian randomization
-
- mRS
-
- modified Rankin Scale
-
- NIHSS
-
- National Institutes of Health Stroke Scale
-
- PCoA
-
- principal coordinates analysis
-
- PERMANOVA
-
- permutational multivariate analysis of variance
-
- rRNA
-
- ribosomal RNA
-
- SAH
-
- S-adenosyl-L-homocysteine
-
- SAM
-
- S-adenosyl-L-methionine
-
- SCFA
-
- short-chain fatty acid
-
- SNV
-
- single nucleotide variant
-
- TOAST
-
- Trial of Org 10172 in Acute Stroke Treatment
-
- TSEA
-
- taxon set enrichment analysis
1 Introduction
Ischemic stroke (IS) is the leading cause of adult disability, with nearly two-thirds of survivors leaving the hospital with impairments [1]. While functional recovery depends on multiple factors, including age, genetics [2], and initial stroke severity [3], a considerable portion of variability in recovery remains unexplained. Various therapeutic approaches—such as the administration of uric acid [4], citicoline [5], and natalizumab [6]—have been explored to restore damaged brain tissue, but none have shown significant benefits. Recently, ApTOLL, a neuroprotective drug targeting toll-like receptor 4, has demonstrated efficacy in patients undergoing endovascular treatment [7].
Emerging evidence suggests that the gut microbiota and its metabolites may play a role in IS risk [8-10]. In ischemia mouse models, the gut microbiota contributes to post-IS variability through bidirectional gut-brain interactions [11]. Microbes are crucial for immune system development and metabolic balance, and disruptions in gut microbiota composition—known as dysbiosis—can lead to pathological effects and increased disease susceptibility [12]. In patients with IS, dysbiosis fosters neuroinflammation [13] due to a rise in opportunistic pathogens and a decline in beneficial bacteria [14]. Additionally, microbial metabolites like short-chain fatty acids (SCFAs) are implicated in central nervous system diseases through immune pathways [15].
Gut microbiota-based therapies offer post-stroke potential for both improving recovery and preventing complications, yet studies on the connection between the gut microbiota and stroke prognosis remain limited. Research in Asian population cohorts has linked unfavorable post-IS recovery outcomes to dysbiosis as characterized by increased pathogenic bacteria and reduced SCFA-producing microbes [16, 17]. However, most such research has relied on small cohorts and used 16S ribosomal RNA (rRNA) gene sequencing, which, compared to the shotgun technique, has limitations regarding detailed species-level and functional profiling.
Hypothesizing that distinct microbial communities and metabolic pathways could influence 3-month post-IS recovery, we applied shotgun metagenomic sequencing to gain insights on the connection between the gut microbiota and stroke prognosis that may refine outcome predictions and guide future therapeutic strategies.
2 Materials and Methods
2.1 Study Design and Participants
Our prospective observational study recruited patients with IS between January 2020 and March 2022 at the Hospital de la Santa Creu i Sant Pau (Barcelona, Spain). Inclusion criteria were age ≥ 18 years and an IS diagnosis confirmed by an expert neurologist based on neuroimaging data.
Excluded were patients who had used antibiotics or probiotics in the month prior to IS onset, patients with recent infections, and patients with a prior disability. Pre-IS functional status was determined by a modified Rankin Scale (mRS) score ≥ 3, as assessed by an mRS-certified stroke neurologist via a simplified dependency questionnaire administered to either the patient or a first-degree relative [S1].
The study adhered to the ethical principles of the Declaration of Helsinki. The hospital's Ethics Committee approved the research protocol (IIBSP-MAE-2020-12), and all participants provided written informed consent to participate.
2.2 Variables
Detailed demographic, clinical, and epidemiological data were prospectively collected from each participant, as follows: age and sex; cardiovascular risk factors (hypertension, dyslipidemia, diabetes mellitus, atrial fibrillation, ischemic heart disease, previous IS, tobacco, and ethanol use); prior medication (antiplatelet agents, oral anticoagulants, statins); stroke etiology as classified according to the Trial of Org 10172 in Acute Stroke Treatment (TOAST) [S2]; baseline and 24-h post-IS neurological deficit severity as assessed using the National Institutes of Health Stroke Scale (NIHSS); type of reperfusion therapy; enema use; and stool consistency evaluated using the Bristol Stool Scale [S3].
On hospital admission, using a structured questionnaire, an mRS-certified stroke neurologist assessed the patient's baseline mRS score (pre-stroke) and 3-month mRS score during outpatient follow-up. Scores of 0–2 and 3–6 were categorized as favorable and unfavorable outcomes, respectively. For individuals unable to attend follow-up in person, the mRS score was assessed in a structured telephone interview.
2.3 Sample Collection
Stool samples collected from each patient following IS onset were processed according to a standardized protocol. The first sample, obtained by a healthcare provider, was immersed in a 96% ethanol solution, and stored at −80°C. Institutional guidelines dictated that oral laxatives be introduced on day 3 of hospitalization if no sample had been collected. If necessary, cleansing enemas were administered from day 5 onwards. As a result, all samples were obtained within five days or less from symptom onset.
2.4 DNA Extraction and Shotgun Sequencing
Stool samples were defrosted and 0.2 g of each was transferred to a 2-mL microcentrifuge tube for DNA extraction using the TIANamp Stool DNA Kit (TIANGEN BIOTECH). Sample concentration was measured with a Qubit fluorometer, and sample integrity and purity were determined with quantitative agarose gel electrophoresis. Genomic DNA was used to construct the libraries, which were pooled and sequenced using Illumina's NovaSeq 6000 System.
2.5 Bioinformatics
Metagenomic DNA sequences were processed and decontaminated from human genome reads using FastQC [S4] v0.11.8. Reads were aligned via BBMap [S5], trimmed, and filtered with Trimmomatic [S6] v0.39. SortMeRNA [S7] v4.2.0 was employed for 16S rRNA sequence extraction, while taxonomic classification was determined using MAPseq [S8] v2.0.1alpha, with the Silva v132 database used for prokaryotic gene assignment.
Microbial sequence read abundance was normalized to account for varying sequencing depths. Paired-end sequences were merged using BBMerge [S5] v38.84, with both merged and unmerged reads assembled into contigs via MegaHit [S9] v1.2.9. Contigs were then annotated using Prokka [S10] v1.14.6, leveraging multiple databases to assign functions to predicted coding sequences.
To quantify functional categories and pathways, original reads were mapped onto assembled contigs using Bowtie2 [S11] v2.3.5. Functional profiles derived from quantified metagenome results enabled comparisons between the favorable and unfavorable post-IS outcome groups. Prokka annotations were used to infer Enzyme Commission (EC) [S12] functions and Kyoto Encyclopedia of Genes and Genomes (KEGG) [S13] pathways.
2.6 Data Filtering and Statistical Analyses
Statistical and bioinformatic analyses were performed using R v4.2.0. Normally distributed and homoscedastic continuous variables were compared using the Student t-test, while non-normally distributed data were analyzed using the Mann–Whitney U-test. Dichotomous variables were evaluated using the chi-squared test.
Gut microbiota diversity was assessed through α-diversity, which evaluates microbial diversity within a single sample, and β-diversity, which compares microbiota composition between samples. α-diversity was estimated using the Shannon, Simpson, observed species, Chao1, and abundance-based coverage estimator (ACE) indexes. The observed species index accounts for the total number of features, while Chao1 and ACE indexes estimate richness (species count). Shannon and Simpson indexes assess both species diversity and distribution evenness. β-diversity was analyzed using principal coordinates analysis (PCoA), with group variation estimated via permutational multivariate analysis of variance (PERMANOVA).
Linear discriminant analysis (LDA) effect size (LEfSe) [S14] was used to identify the species that differed significantly between the two outcome groups; a logarithmic LDA score ≥ 2 and an adjusted p < 0.05 were considered significant. The LEfSe algorithm employs the Kruskal–Wallis rank sum test to detect bacteria with differential abundance, while LDA evaluates their relevance.
A linear model, adjusted for age, sex, IS subtype (TOAST classification), and stroke severity (NIHSS), was used to discriminate taxa and metabolic pathways that differed significantly between the two outcome groups. These clinical variables, selected via logistic regression, analyzed variable relationships with 3-month outcomes (Table S1).
To identify disease-associated taxonomic signatures, we used Taxon Set Enrichment Analysis (TSEA) via the MicrobiomeAnalyst module [S15], a variation of Gene Set Enrichment Analysis (GSEA) adapted for microbiome sequencing data. Similar to GSEA, TSEA performs hypergeometric tests against a taxon set library to identify relevant microbial signatures.
Sequencing errors and low-level contaminants were managed by removing rare taxa, defined as those present in < 25% of samples [S16]. To mitigate false-positive associations from multiple testing, p-values were adjusted using the Bonferroni correction (adjusted p < 0.05).
2.7 Mendelian Randomization Analyses
To evaluate potential causal relationships between significant gut bacteria, metabolic pathways, and post-IS outcomes, Mendelian randomization (MR) analyses were conducted using publicly available genome-wide association study (GWAS) summary statistics (Table S2). Exposure GWAS included abundance data for multiple bacterial taxa (Pseudomonadales [S16], Enterococcaceae [S16], Enterococcus [S16], Bacteroides fragilis [S17], Faecalibacterium prausnitzii [S17], Paraprevotella clara [S17], Sutterella wadsworthensis [S17], Eubacterium eligens [S17]), as well as pyruvate [S18] and S-adenosyl-L-homocysteine [S19] (SAH). Outcome GWAS data were sourced from GISCOME [S20], a meta-analysis of 12 IS recovery cohorts (6165 cases from across Europe, the USA, and Australia) that evaluated functional status 60–190 days post-IS using mRS dichotomous scoring (0–2 vs. 3–6), adjusted for age, sex, and IS severity.
MR analyses were conducted using the TwoSampleMR [S21] package. Summary statistics were filtered (p < 5 × 10−6) and clumped using the European 1000 Genomes Project reference panel (r2 < 0.01, clump distance > 10,000 kb) [S22]. Ambiguous and palindromic single nucleotide variants (SNVs) were excluded during harmonization.
MR methods applied were inverse-variance weighted (IVW), MR-Egger, weighted median, penalized weighted median, and weighted mode, with IVW serving as the primary approach. Horizontal pleiotropy and heterogeneity were assessed using Egger regression [S23] and the Cochran Q statistic [S24], respectively. To detect potential outliers influencing results, leave-one-out analysis was performed for significant MR results.
The minimum causal relationship required for 80% MR power (α = 0.05) was estimated using Burgess' online calculator, considering sample size conditions and the proportion of variance explained by the instruments [S25].
2.8 Reporting Guidelines
This article follows the Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) and STROBE-MR [S26] reporting guidelines.
3 Results
3.1 Study Participants
Of a total of 731 eligible subjects, metagenomic sequencing was performed on fecal samples from 128 patients with IS, 84 (65.6%) with favorable and 44 (34.4%) with unfavorable 3-month functional outcomes (Figure S1). Stool samples were collected between days 1 and 4 in 82% of patients.
Patient baseline characteristics are reported in Table 1. Regarding variability between the two outcome groups, patients with unfavorable outcomes were older, were more likely to have a history of hypertension, experienced greater stroke severity, and more frequently underwent mechanical thrombectomy procedures; they were also more frequently administered enemas, which would explain stool consistency differences found between the two groups.
| Total (n = 128) | mRS 0–2 (n = 84, 65.6%) | mRS 3–6 (n = 44, 34.4%) | p | |
|---|---|---|---|---|
| Sex, (%) | ||||
| Male | 77 (60.2) | 53 (63.1) | 24 (54.5) | 0.348 |
| Demographics, median (IQR) | ||||
| Age, years | 76.0 (63.5–82) | 72.5 (59–79.3) | 80.5 (73.8–86) | < 0.001* |
| Height, cm | 167.0 (157.5–172) | 167.0 (157.3–172) | 165.0 (158–171) | 0.929 |
| Weight, kg | 73.5 (65–79.8) | 73.5 (65.1–79.5) | 73.5 (64–80) | 0.759 |
| BMI, kg/m2 | 26.0 (24.2–28.5) | 25.9 (24.1–28.4) | 26.1 (24.8–28.5) | 0.605 |
| Risk factors, n (%) | ||||
| Hypertension | 90 (70.3) | 53 (63.1) | 37 (84.1) | 0.014* |
| Dyslipidaemia | 73 (57.0) | 46 (54.8) | 27 (61.4) | 0.474 |
| Diabetes mellitus | 29 (22.7) | 17 (20.2) | 12 (27.3) | 0.367 |
| Atrial fibrillation | 30 (23.4) | 16 (19.0) | 14 (31.8) | 0.105 |
| Ischemic heart disease | 21 (16.4) | 14 (16.7) | 7 (15.9) | 0.913 |
| Previous ischemic stroke | 21 (16.4) | 14 (16.7) | 7 (15.9) | 0.913 |
| Tobacco use | 22 (17.2) | 17 (20.2) | 5 (11.4) | 0.206 |
| Ethanol use | 33 (25.8) | 24 (28.6) | 9 (20.4) | 0.319 |
| Pre-stroke medication, n (%) | ||||
| Antiplatelet agents | 28 (21.9) | 17 (20.2) | 11 (11.4) | 0.536 |
| Oral anticoagulants | 18 (14.1) | 11 (13.1) | 7 (15.9) | 0.664 |
| Statins | 47 (36.7) | 33 (39.3) | 14 (31.8) | 0.405 |
| Reperfusion therapy, n (%) | ||||
| IV tPA | 41 (32.0) | 28 (33.3) | 13 (29.5) | 0.663 |
| Mechanical thrombectomy | 35 (27.3) | 18 (21.4) | 17 (38.6) | 0.014* |
| NIHSS score, median (IQR) | ||||
| Baseline | 5.0 (3–12) | 4.0 (2–9) | 8.5 (4.8–16.3) | < 0.001* |
| 24-h | 3.0 (1–7) | 2.0 (0–4) | 7.0 (3–15) | < 0.001* |
| TOAST stroke subtype, n (%) | ||||
| Cardioembolism | 35 (27.3) | 22 (31.2) | 13 (29.5) | 0.686 |
| Large artery atherosclerosis | 23 (18.0) | 15 (17.9) | 8 (18.2) | 0.964 |
| Small vessel disease | 12 (9.4) | 7 (8.3) | 5 (11.4) | 0.811 |
| Other etiology | 13 (10.2) | 11 (13.1) | 2 (4.5) | 0.225 |
| Undetermined | 45 (35.2) | 29 (34.5) | 16 (36.4) | 0.990 |
| Blood laboratory findings, median (IQR) | ||||
| Glucose, mg/100 mL | 113.4 (99.5–147.9) | 112.7 (97.3–133.3) | 122.1 (102.4–163.8) | 0.096 |
| White blood cell count, ×109/L | 8.2 (6.8–10.1) | 7.8 (6.9–9.6) | 8.8 (6.7–10.7) | 0.172 |
| Enema use and Bristol Stool Scale score, n (%) | ||||
| Enema | 23 (18.0) | 9 (10.7) | 14 (31.8) | 0.003* |
| Type 1–2 (constipation) | 27 (21.1) | 23 (27.4) | 4 (9.1) | 0.016* |
| Type 3–4 (normal) | 61 (47.7) | 43 (51.2) | 18 (40.9) | 0.269 |
| Type 5 (lacking fiber) | 17 (13.3) | 8 (9.5) | 9 (20.4) | 0.084 |
| Type 6–7 (diarrhea) | 23 (18.0) | 10 (11.9) | 13 (29.5) | 0.014* |
- Abbreviations: BMI, body mass index; IQR, interquartile range; IV tPA, intravenous tissue-type plasminogen activator; mRS, modified Rankin Scale; NIHSS, National Institutes of Health Stroke Scale; TOAST, Trial of Org 10172 in Acute Stroke Treatment.
- * p < 0.05.
3.2 Gut Microbiota Diversity
We identified 46 classes, 78 orders, 131 families, 279 genera, and 240 species, whose distributions were measured within the samples. Class-level α-diversity analyses of the microbial community showed that bacterial richness (observed species index, pclass = 0.017; Chao1 index, pclass = 0.026; ACE index, pclass = 0.017) was statistically higher in patients with favorable 3-month post-IS outcomes (Figure 1A). For orders, families, genera, and species, however, α-diversity differences were not statistically significant (Table S3).
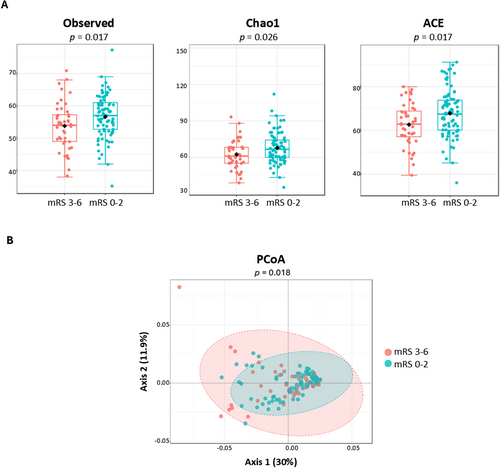
Regarding taxa distribution in the samples, we plotted PCoA based on Bray-Curtis dissimilarities and measured the β-diversity using PERMANOVA. A strong separation was observed in microbial structure at the family (pfamily = 0.030), genus (pgenus = 0.035), and species (pspecies = 0.018) levels between patients with favorable and unfavorable outcomes (Figure 1B).
3.3 Gut Microbiota Taxonomic Differences
Analysis using LEfSe revealed that the unfavorable outcomes group had a greater abundance of pathogenic bacteria, including Enterobacter sp. UPIB, Erysipelotrichaceae bacterium 6_1_45, Clostridium clostridioforme, Clostridium bolteae, Bacteroides fragilis, Eisenbergiella tayi, Clostridium symbiosum, Hungatella hathewayi, and Neglecta timonensis, as well as a lesser abundance of the commensal bacteria Faecalibacterium prausnitzii, Paraprevotella clara, Blautia stercoris, Sutterella wadsworthensis, Roseburia faecis, Coprobacillus sp. 8_1_38FAA, and Eubacterium eligens (Figure 2A, Table S4).
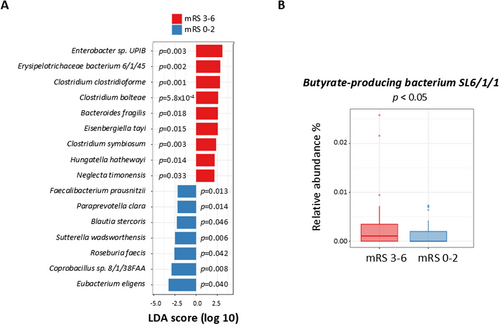
Further statistical analysis adjusting for age, sex, stroke severity, and stroke subtype revealed different relative taxa abundance between the two outcome groups (Table 2). For the unfavorable outcomes group, Synergistia was increased at the class level; Synergistales and Pseudomonadales were increased at the order level; Synergistaceae, Pseudomonadaceae, and Enterococcaceae were increased at the family level; Pseudomonas, Finegoldia, Porphyromonas, and Enterococcus were increased at the genus level; and finally, Butyrate-producing bacterium SL6/1/1 was increased at the species level (Figure 2B). TSEA, using the different relative taxa abundance between the two groups, showed an association between our taxon set library and stroke (p = 0.036). Nevertheless, the MR analysis did not point to any causal relationship between these bacteria and post-IS functional outcomes (Table S5). MR results were not biased by heterogeneity nor pleiotropy (Table S6), although MR power calculations suggested that the MR analysis was underpowered (Table S7).
| Mean relative abundance ± SE (%) | p | β | 95% CI | ||
|---|---|---|---|---|---|
| mRS 0–2 | mRS 3–6 | ||||
| Phylum | |||||
| Synergistetes | 0.051 ± 0.014 | 0.139 ± 0.046 | 0.002 | −1.113 | −1.756, −0.498 |
| Acidobacteria | 0.036 ± 0.006 | 0.130 ± 0.039 | 0.008 | −0.849 | −1.382, −0.333 |
| Class | |||||
| Synergistia | 0.046 ± 0.012 | 0.132 ± 0.045 | 0.001 | −1.127 | −1.756, −0.524 |
| Order | |||||
| Synergistales | 0.046 ± 0.012 | 0.133 ± 0.045 | 0.002 | −1.134 | −1.765, −0.528 |
| Pseudomonadales | 0.187 ± 0.013 | 0.362 ± 0.066 | 0.012 | −0.519 | −0.804, −0.239 |
| Family | |||||
| Synergistaceae | 0.046 ± 0.013 | 0.137 ± 0.047 | 0.005 | −1.137 | −1.777, −0.524 |
| Pseudomonadaceae | 0.118 ± 0.011 | 0.272 ± 0.063 | 0.006 | −0.674 | −1.025, −0.330 |
| Enterococcaceae | 0.089 ± 0.016 | 0.407 ± 0.171 | 0.052 | −1.103 | −1.815, −0.431 |
| Genus | |||||
| Pseudomonas | 0.095 ± 0.011 | 0.292 ± 0.086 | 0.002 | −0.859 | −1.276, −0.453 |
| Finegoldia | 0.004 ± 0.001 | 0.013 ± 0.005 | 0.016 | −1.744 | −2.827, −0.670 |
| Porphyromonas | 0.005 ± 0.001 | 0.033 ± 0.015 | 0.032 | −1.242 | −2.038, −0.499 |
| Enterococcus | 0.069 ± 0.015 | 0.425 ± 0.189 | 0.054 | −1.200 | −1.966, −0.480 |
| Species | |||||
| Butyrate-producing bacterium SL6/1/1 | 0.001 ± 0.000 | 0.003 ± 0.001 | 0.026 | −1.098 | −1.668, −0.543 |
- Abbreviations: β, mean change in the dependent variable given a one-unit change in the independent variable; CI, confidence interval; IQR, interquartile range; mRS, modified Rankin Scale; SE, standard error.
3.4 Gut Microbiota Functional Differences
Mapping of metagenomic sequences against the KEGG pathway [S13] database revealed functional profiles for the samples. A total of 165 KEGG pathways were annotated, revealing significant differences between the outcome groups in the ethylbenzene degradation pathway (map00642), which was increased in the unfavorable outcomes group (p = 4.9 × 10−2, β = −0.561, 95% CI: −0.893, −0.237) (Figure 3A).
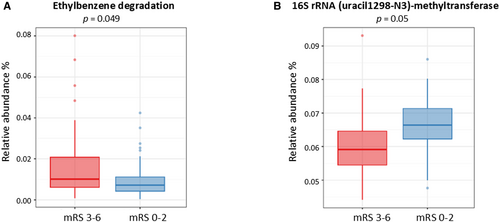
Metagenomic sequences were also mapped against the EC Number [S12] database and classified into 1225 genes. Enzyme 2.1.1.193, also known as 16S rRNA (uracil1498-N3)-methyltransferase, was reduced in the unfavorable outcomes group (p = 5.0 × 10−2, β = 0.101, 95% CI: 0.052, 0.150) (Figure 3B).
The MR analysis showed a causal relationship between pyruvate, one of the products of ethylbenzene degradation, and post-IS functional outcomes (pIVW = 0.014, β = −0.502). Furthermore, although it did not reach significance, the MR analysis pointed to an association between levels of SAH, as the main product of 16S rRNA (uracil1498-N3)-methyltransferase, and post-IS functional outcomes (pIVW = 0.150, β = 0.182) (Table 3). MR results were not biased by heterogeneity, pleiotropy, or outliers (Table S8).
| Exposure | Outcome | Method | SNVs | p | β | OR (95% CI) |
|---|---|---|---|---|---|---|
| Pyruvate levels | GISCOME | IVW | 16 | 0.014* | −0.502 | 0.605 (0.404–0.905) |
| MR-Egger | 16 | 0.736 | −0.103 | 0.901 (0.499–1.628) | ||
| Weighted median | 16 | 0.401 | −0.204 | 0.814 (0.504–1.315) | ||
| Penalized weighted median | 16 | 0.434 | −0.202 | 0.816 (0.491–1.357) | ||
| Weighted mode | 16 | 0.320 | −0.229 | 0.794 (0.512–1.232) | ||
| S-adenosyl-L-homocysteine levels | GISCOME | IVW | 15 | 0.150 | 0.182 | 1.200 (0.936–1.538) |
| MR-Egger | 15 | 0.186 | 0.428 | 1.534 (0.840–2.802) | ||
| Weighted median | 15 | 0.268 | 0.195 | 1.216 (0.859–1.721) | ||
| Penalized weighted median | 15 | 0.275 | 0.195 | 1.216 (0.855–1.729) | ||
| Weighted mode | 15 | 0.356 | 0.212 | 1.236 (0.799–1.914) |
- Abbreviations: β, estimation of the causal relationship between exposure and outcome; CI, confidence interval; GISCOME, Genetics of Ischemic Stroke Functional Outcome; IVW, inverse variance weighted; MR, Mendelian randomization; OR, odds ratio; SNV, single nucleotide variant.
- * p < 0.05.
4 Discussion
In this study, we analyzed variations in gut microbiota in patients with IS categorized into favorable and unfavorable 3-month functional outcome groups. In the microbial communities of patients with unfavorable compared to favorable outcomes, α-diversity analysis showed comparatively reduced bacterial richness, while β-diversity analysis revealed structural differences characterized by an increase in opportunistic pathogens and a decrease in bacteria associated with positive recovery. Significant differences were also observed between the two groups in metabolic pathways and enzyme activity.
Certain factors directly influenced prognosis for the two groups, such as baseline NIHSS scores and the frequency of mechanical thrombectomy procedures. Additionally, more frequent in the unfavorable outcome group was enema use, which may affect gut microbiota composition [18], while constipation is a common post-IS complication linked to poorer neurological outcomes, increased complications, and even mortality [19].
Since gut microbiota composition and stroke outcomes are influenced by factors such as age and sex, we adjusted for these variables in our analyses [20]. Also accounted for was baseline neurological severity (evaluated using the NIHSS), as it is the most reliable predictor of 3-month outcomes [21]. Stroke etiology was likewise taken into account, as certain subtypes (e.g., cardioembolic stroke) are associated with poorer outcomes. Although we pre-specified these confounding factors to minimize bias, residual confounding may still exist.
The gut microbiota plays a fundamental role in several physiological processes, including metabolism, nutrient absorption, neurotransmitter regulation, barrier integrity, and immune system modulation [22], and dysbiosis is widely recognized as detrimental to microbial composition. A key hallmark of gut dysbiosis is reduced microbiota diversity [23], which has been linked to neurological disorders via the gut-brain axis [24]. Our observation of reduced α-diversity in patients with unfavorable outcomes aligns with findings reported for Chinese patients with IS [17]. Gut microbiota diversity has been associated with blood–brain barrier integrity, permeability, and potential leakage [14]. Dysbiosis can prompt immune cell recruitment to the central nervous system, contributing to neuroinflammation [22]—a critical factor in brain damage. Pro-inflammatory cytokines facilitate further leukocyte recruitment from peripheral blood [14], exacerbating neuronal cell death, enlarging the infarcted area, and leading to more severe neurological outcomes [19, 25-28].
Using LEfSe analysis [S14], we identified a set of bacteria that were likely to contribute to differences between patients with favorable and unfavorable 3-month functional outcomes. Patients with unfavorable outcomes showed a greater abundance of several pathogenic bacteria, including Enterobacter spp., Erysipelotrichaceae spp., Clostridium clostridioforme, Clostridium bolteae, Bacteroides fragilis, Eisenbergiella tayi, Clostridium symbiosum, Hungatella hathewayi, and Neglecta timonensis, and conversely, showed a lower abundance of commensal bacteria such as Faecalibacterium prausnitzii, Paraprevotella clara, Blautia stercoris, Sutterella wadsworthensis, Roseburia faecis, Coprobacillus spp., and Eubacterium eligens. Our findings align with those of previous studies on Asian populations, despite differences in sequencing techniques [16, 17]. In patients with favorable outcomes, Chang et al. [16] reported increased Blautia levels, matching our observation of higher Blautia stercoris abundance. In patients with unfavorable outcomes, Sun et al. [17] found lower Faecalibacterium and increased Clostridium innocuum and Erysipelotrichaceae levels, similar to our finding of decreased Faecalibacterium prausnitzii and elevated Clostridium bolteae, Clostridium clostridioforme, Clostridium symbiosum, and Erysipelotrichaceae spp. levels.
After adjusting for the variables associated with 3-month outcomes, we continued to observe significant differences in bacterial taxa between the favorable and unfavorable outcome groups. In the unfavorable outcomes group, there was a greater abundance of Synergistia at the class level, Synergistales and Pseudomonadales at the order level, Synergistaceae, Pseudomonadaceae, and Enterococcaceae at the family level, Pseudomonas, Finegoldia, Porphyromonas, and Enterococcus at the genus level, and Butyrate-producing bacterium SL6/1/1 at the species level. Enrichment analysis using TSEA showed an association between our list of significant taxa and stroke, suggesting that our specific set of bacteria is enriched in patients with IS.
Our findings suggest an increased presence of opportunistic pathogens in patients with unfavorable outcomes. Elevated Acidobacteria levels have been linked to acute coronary syndrome [29] and Alzheimer's disease [30], while Synergistetes is associated with various pathological conditions; higher Synergistaceae abundance is observed, for instance, in Parkinson's disease. Certain Pseudomonas species have been implicated in both ischemic and haemorrhagic cerebrovascular diseases [31]. Porphyromonas may function as either a key player in host immunity or a pathobiont that triggers disease when homeostasis is disrupted [32]. Additionally, Enterococcaceae and its most abundant genus, Enterococcus, are positively correlated with high-risk factors in patients with IS [33]. Clinical studies have consistently shown that these patients exhibit an altered gut microbiota profile, marked by increased pathogenic bacteria and reduced probiotics [34-36]. However, further research is needed to better understand microbial dysbiosis patterns and the specific role played by those bacteria in 3-month post-IS outcomes.
Given that the gut-brain axis is a complex bidirectional network linking the intestine and the central nervous system, we performed MR analysis using GWAS data (GISCOME dataset) to assess whether bacterial abundance changes were causally associated with 3-month unfavorable outcomes. However, we found no evidence to support a causal link. Given significant interindividual variability and microbiota heterogeneity, microbiota GWAS analyses often suffer from reduced statistical power [37]. To address this issue, we conducted a power analysis, which revealed that our conditions were insufficient to accurately detect genetic effects, requiring a stronger causal relationship between gut microbiota and post-IS recovery.
Our study revealed metabolic differences between patients with favorable and unfavorable outcomes. The unfavorable group exhibited an increased abundance of the ethylbenzene degradation pathway, affecting metabolites—like pyruvate—that play roles in other metabolic processes. The fact that ethylbenzene has been linked to oxidative damage and apoptosis in rat brain tissue [38] suggests a potential negative impact on stroke recovery. However, degradation products like pyruvate offer therapeutic promise, as they have been shown to reduce infarct volume in ischemia mouse models [39]. Our MR analysis confirmed a causal relationship between pyruvate and post-IS functional outcomes, with higher pyruvate levels correlating with favorable recovery. Conversely, found to be less abundant in the unfavorable outcomes group was 16S rRNA (uracil1498-N3)-methyltransferase, an enzyme that catalyzes the conversion of S-adenosyl-L-methionine (SAM) to SAH. While SAM supplementation has demonstrated benefits for ischemia recovery in animal models [40], elevated SAH levels have been linked to poorer stroke outcomes and increased mortality risk [40-42]. All in all, those findings highlight metabolic pathway differences between favorable and unfavorable outcomes, with potential implications for ethylbenzene metabolism and SAM/SAH regulation in IS recovery. However, this association is based on cross-sectional data, and information on microbiota evolution over time remains scarce. We hypothesize that ethylbenzene degradation and pro-inflammatory pathways may influence this process. To analyze the functional profile while avoiding statistical overadjustment, we adjusted for established prognostic factors while limiting our model to age, sex, stroke etiology, and stroke severity.
In conclusion, our study highlights key differences in gut microbiota composition in patients with unfavorable versus favorable 3-month post-IS outcomes, namely, reduced diversity in bacteria associated with positive recovery, increased levels of opportunistic pathogens, a greater abundance of the ethylbenzene degradation pathway, and lower 16S rRNA (uracil1498-N3)-methyltransferase enzyme activity.
Our findings suggestive of a potential link between gut microbiota and long-term post-IS outcomes implicate microbial alterations in the pathogenesis and progression of IS-related complications. Therefore, targeting gut microbiota modulation may offer a promising therapeutic avenue for post-IS treatment. However, further research is necessary to elucidate the underlying mechanisms and evaluate the safety and efficacy of such interventions.
4.1 Study Strengths and Limitations
This study leveraging shotgun metagenomic sequencing to identify microbiota signatures linked to 3-month post-IS outcomes has several limitations. Our findings may lack broad generalizability given that the study was conducted in a single center, although this ensured a more homogeneous sample with reduced external variability. Patient groups differed, although bias was mitigated by adjusting for multiple clinical variables. Notably, the proportion of patients undergoing mechanical thrombectomy—an important prognostic factor—varied between groups. Dietary and physical activity data were not collected, although hospitalized patients received similar diets, and the first fecal sample was collected at stroke onset, meaning that microbiota profiles may have been altered as time passed. A limitation that we plan to address in future studies was that we could not conduct additional assays or metabolomic profiling due to budget constraints. Despite adjusted analyses, baseline differences—such as vascular risk factors—may limit results validity. Finally, as an observational study, causal links between microbiota and 3-month functional outcomes could not be established, and although we identified relevant associations, the statistical power for the MR analysis was insufficient. To refine understanding of the gut microbiota's role in IS recovery, future research should focus on larger sample sizes, more comprehensive patient data, and mechanistic investigations.
Author Contributions
Miquel Lledós: investigation, writing – original draft, writing – review and editing, methodology, visualization, formal analysis. Luís Prats-Sánchez: writing – review and editing, writing – original draft, investigation. Elena Muiño: writing – review and editing. Natalia Cullell: writing – review and editing. Laia Llucià-Carol: writing – review and editing. Jara Cárcel-Márquez: writing – review and editing. Cristina Gallego-Fabrega: writing – review and editing. Jesús M. Martín-Campos: writing – review and editing. Rebeca Marín: writing – review and editing. Ana Aguilera-Simón: writing – review and editing. Marina Guasch-Jiménez: writing – review and editing. Garbiñe Ezcurra-Díaz: writing – review and editing. Pol Camps-Renom: writing – review and editing. María del Mar Freijo: writing – review and editing. Joan Martí-Fàbregas: writing – review and editing. Israel Fernández-Cadenas: funding acquisition, supervision, resources, writing – review and editing, conceptualization.
Acknowledgments
We thank all patients and staff who participated in this research. Special thanks to the nursing team (see all study participants in the Supporting Information).
Conflicts of Interest
The authors declare no conflicts of interest.
Open Research
Data Availability Statement
Study data are available on request from the corresponding author.




