Autophagic compound database: A resource connecting autophagy-modulating compounds, their potential targets and relevant diseases
Abstract
Objectives
Autophagy, a highly conserved lysosomal degradation process in eukaryotic cells, can digest long-lived proteins and damaged organelles through vesicular trafficking pathways. Nowadays, mechanisms of autophagy have been gradually elucidated and thus the discovery of small-molecule drugs targeting autophagy has always been drawing much attention. So far, some autophagy-related web servers have been available online to facilitate scientists to obtain the information relevant to autophagy conveniently, such as HADb, CTLPScanner, iLIR server and ncRDeathDB. However, to the best of our knowledge, there is not any web server available about the autophagy-modulating compounds.
Methods
According to published articles, all the compounds and their relations with autophagy were anatomized. Subsequently, an online Autophagic Compound Database (ACDB) (http://www.acdbliulab.com/) was constructed, which contained information of 357 compounds with 164 corresponding signalling pathways and potential targets in different diseases.
Results
We achieved a great deal of information of autophagy-modulating compounds, including compounds, targets/pathways and diseases. ACDB is a valuable resource for users to access to more than 300 curated small-molecule compounds correlated with autophagy.
Conclusions
Autophagic compound database will facilitate to the discovery of more novel therapeutic drugs in the near future.
1 INTRODUCTION
Autophagy, the well-known intracellular process of degrading or recycling damaged organelles, long-lived proteins and other cellular components, can maintain homeostasis and energy balance in cells.1 This term is derived from the ancient Greek for “self-eating”, named by Christian de Duve in 1963.2 Depending on the transportation modes of intracellular substrates to lysosomes, autophagy can be divided into 3 identified types, microautophagy, chaperone-mediated autophagy and macroautophagy (unless stated, macroautophagy is referred to as autophagy).3 Recent studies have demonstrated that dysregulation of autophagy is involved in several pathological processes, such as cancer, cardiovascular disorders, diabetes, neurodegeneration and ageing.4-8 Many autophagy-related proteins have been proved as therapeutic targets such as ULK1 (Unc51-like protein kinase 1), Beclin-1, ubiquitin-like-conjugating enzyme Atg3, AMPK (adenosine 5′-monophosphate (AMP)-activated protein kinase) and so on.9-12 Therefore, the regulation of autophagy will become an important therapeutic and diagnostic research field.
In 2016, Yoshinori Ohsumi received the Nobel Prize in physiology or medicine for his contribution to the discovery of autophagy mechanisms. Because of the efforts of Ohsumi and other scientists, we know that autophagy controls some important physiological mechanisms. Nowadays, mechanisms of autophagy have been gradually elucidated and thus targeting autophagy for small-molecule drug discovery has been receiving a rising attention.13, 14 In recent years, there is growing evidence that small-molecule compounds could regulate the progression of autophagy and displayed great therapeutic potential in various diseases. Tert-butylhydroquinone (tBHQ) could protect hepatocytes against saturated fatty acids induced-lipotoxicity, via AMPK-dependent autophagy induction.15 Hepatocellular carcinoma has high rate of fatality; galangin could induce autophagy by activating AMPK and suppressing mTOR to function as anti-proliferation effect in vitro and in vivo.16 In neurodegenerative diseases, rilmenidine administration relieved signs and decreased levels of Huntington's disease in vivo, which was partially due to its effect on enhancement of autophagy.17 The neuroprotection of deferoxamine on Parkinson's disease was also contributed to activation of HIF-1α and HIF-1α-mediated induction of autophagy.18 Moreover, Lu AE58054 and nicotinamide could directly or indirectly induce cytoprotective autophagy in Alzheimer's disease, which were undergoing clinical trials in different phases.19, 20
Currently, some autophagic web servers have been available online to facilitate scientists to query autophagic resources conveniently, such as HADb (Human autophagy database), CTLPScanner (Chromothripsis-like pattern scanner), iLIR server, and ncRDeathDB (ncRNA-associated cell death database).21-24 However, to the best of our knowledge, there is not any web server available about autophagy-modulating compounds. Thus, in this study, an online Autophagic Compound Database (ACDB) (http://www.acdbliulab.com/) was constructed, which contained information of compounds with their targets and relevant signalling pathways in diseases.
2 MATERIALS AND METHODS
2.1 Web server generation
Information of ACDB web server was developed on Linux by using MySQL (version 5.1.48-log, Oracle Corporation, Redwood Shores, California, USA) database management system. ACDB web interface was built on web technologies, including PHP (versions 7.1, Zend Technologies, Cupertino, California, USA), CSS (cascading style sheets), HTML (hypertext markup language) and JavaScript (Oracle Corporation, Redwood Shores, California, USA). Apache (version 2.4, The Apache Software Foundation, Wakefield, Massachusetts, USA) web server technology was used to develop and serve web application.
2.2 Web server construction and structure
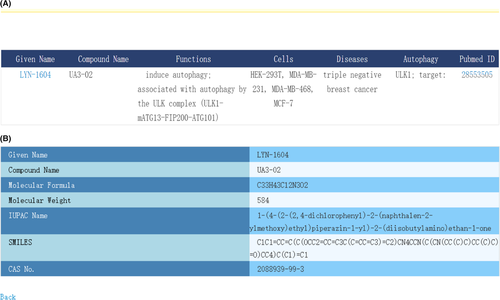
Autophagic compound database was composed of 2 primary data sheets, namely autophagy-related information (sheet 1) and compound (sheet 2). The autophagy-related information was divided into 7 parts, named No., PubMed ID, title, function, cell, disease name, pathway/potential target, respectively. We searched for all compounds and their relations with autophagy by text-mining method from related databases and published articles. The PubMed Unique Identifiers (PMIDs) and titles were collected as contents of PubMed ID and title fields. Information of autophagy-modulating compounds was inputted into the function field. The data fields such as cell, disease name, pathway/potential target were filled by authors mentioned in the article. There were 8 fields in compound sheet, including No., given name, compound name, CAS No., molecular formula, molecular weight, IUPAC name and SMILES expression. IUPAC name represented the International Union of Pure and Applied Chemistry name, and SMILES expression meant Simplified Molecular Input Line Entry Specification expression of the specific compound. “No.” as the same field name of 2 sheets was performed as the connection between compound and autophagy-related information.
2.3 Literature information collection
Precise information of sheet 1 was collected from related public databases and text-mining of published articles manually. PubMed ID was linked to the relevant webpage. The compound name, CAS No. and IUPAC name in compound sheet were mainly integrated and verified by Scifinder Scholar (https://scifinder.cas.org), ChemcalBook (http://www.chemicalbook.com) and Reaxys (http://new.reaxys.com). The software ChemBioDraw Ultra 14.0.0.117 was used to draw the structure and confirm the molecular formula and weight. SMILES expressions of compounds were converted by their hand-painted structures. All the details were checked and verified by the public databases to ensure their accuracy.
3 RESULTS
3.1 The extracted information of ACDB
In general, we searched for all compounds and their relations with autophagy by text-mining method from published articles. Precise information of compounds was mainly integrated by Scifinder Scholar (https://scifinder.cas.org), ChemcalBook (http://www.chemicalbook.com) and Reaxys (http://new.reaxys.com). In total, 357 compounds that regulated autophagic pathways were extracted from 457 references (Table 1). Moreover, we collected and normalized the related information of these compounds, including Consummated Chemical Abstracts Service Registry Number (CAS No.), molecular formula, molecular weight and International Union of Pure and Applied Chemistry (IUPAC) name. All these information were collected from public databases or related documents. Simplified Molecular Input Line Entry Specification (SMILES) expressions of compounds were converted by their hand-painted structures in ChemBioDraw Ultra. To obtain the functional constituents of these compounds, we manually investigated related articles for data of cells, potential targeted proteins, signalling pathways and diseases.
| Data field | Data source | Amount of data |
|---|---|---|
| Compounds | ChemicalBook, SciFinder, Reaxys, PubMed | 357 |
| Pathways | PubMed, text-mining | 96 |
| Potential targets | PubMed, text-mining | 68 |
| Cells types | PubMed, text-mining | 443 |
Moreover, ACDB integrated data about the effects of compounds on autophagy and related mechanisms. Among them, 298 compounds could directly induce autophagy, and 43 compounds could inhibit autophagy. Furthermore, 16 compounds such as AU0119 (melatonin), AU0232 (chloroquine) and AU0255 (berberine) showed different functions of inducing or inhibiting autophagy in specific cells or diseases. Sixty-three compounds were discovered to have potential targets and 83 compounds were found to influence corresponding signalling pathways. AU0254 (beta-asarone) and AU0262 (AZD8055) could regulate autophagy by different pathways or receptor proteins in different diseases. For example, AU0254 functions neuroprotective effect in Parkinson's disease through JNK/Bcl-2/Beclin-1 pathway, whereas in Alzheimer's disease model, it protected cells via activation of Akt/mTOR pathway.
3.2 Web server query
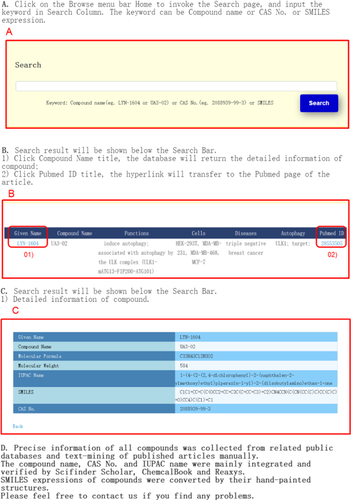
We generated a friendly web interface for users to easily explore the web server. Users can obtain results via fuzzy search, with compound name, CAS No. or SMILES expression as keywords (Figure 1). The result page can display the information of the compound and its connection with autophagy (Figure 2A). Users can also follow the provided hyperlinks to obtain detailed information (Figure 2B). In addition, a help hyperlink on the website is provided as an instruction to users (Figure 3). For instance, users can search by LYN-1604 (compound name), 2088939-99-3 (CAS No.) or its SMILES expression and other search-related index term to obtain the information of this compound and related functions. In this search result, ULK1 is the target protein of LYN-1604 which could induce autophagy. We found that LYN-1604 has therapeutic potential to trip-negative breast cancer by targeting ULK1.9 Here, cell names were also presented, such as HEK-293T, MDA-MB-231, MDA-MB-468 and MCF-7 cells. The diseases associated with the study or that authors mentioned in articles were also provided. In addition, the PMIDs and titles of all articles were provided for ACDB users to obtain detailed information.
4 DISCUSSION
Autophagy refers to the natural and regulated mechanism of normal cells that disposes aged and damaged cellular components. Accumulating researches have indicated that autophagy may possess contradictory functions in certain conditions.25 Depending on our data collation, small-molecule compounds could induce or inhibit autophagy via different signal pathways or targets in certain diseases.26-29 Recently, plenty of researches have proved that autophagy-modulating compounds may have noteworthy therapeutic effects on cancers, neurodegenerative diseases and so on.30, 31 The design and development of small-molecule compounds targeting autophagy will be a promising therapeutic approach for treatment or diagnosis of diseases. However, few compounds developed as autophagic medicines have been approved for clinical use or in clinical trial. Consequently, ACDB could provide an avenue for rapidly querying and estimating autophagic-activated or -inhibited compounds for users.
In conclusion, 357 compounds, 68 potential targets, 96 signalling pathways and 443 types of cells that related to autophagy have been included in ACDB. The online interface allows users to query compounds for related targets, signalling pathways and cells and diseases. It can also help users to estimate targets and relevant pathways of synthetic or isolated compounds based on structural similarities. ACDB will be updated regularly to facilitate the research of autophagy. We believe that ACDB is a valuable resource for users to access to more than 300 curated compounds correlated to autophagy, contributing to the discovery of novel therapeutic targets in the future. Moreover, ACDB will facilitate to the discovery of more novel therapeutic drugs in the near future.
ACKNOWLEDGEMENTS
This work was supported by grants from National Key R&D Program (2017YFC0909302), and National Natural Science Foundation of China (Grant No. 81673455 and 81402496).
CONFLICT OF INTEREST
The authors declare no conflict of interest.