Trends & Opportunities in Visualization for Physiology: A Multiscale Overview
Abstract
Combining elements of biology, chemistry, physics, and medicine, the science of human physiology is complex and multifaceted. In this report, we offer a broad and multiscale perspective on key developments and challenges in visualization for physiology. Our literature search process combined standard methods with a state-of-the-art visual analysis search tool to identify surveys and representative individual approaches for physiology. Our resulting taxonomy sorts literature on two levels. The first level categorizes literature according to organizational complexity and ranges from molecule to organ. A second level identifies any of three high-level visualization tasks within a given work: exploration, analysis, and communication. The findings of this report may be used by visualization researchers to understand the overarching trends, challenges, and opportunities in visualization for physiology and to provide a foundation for discussion and future research directions in this area.
1. Introduction
Human physiology describes the functions and mechanisms of the human body that make it a living being. Forming the link between the basic sciences (biology, chemistry, and physics) and medicine, human physiology is multiscale in that it integrates the individual functions of molecules, cells, tissues, and organs into a whole organism [HG11]. Physiology is an important aspect of systems biology, which has been characterized as an approach to understanding multiscale interactions in a biological system [KN09]. While systems biology tends toward data-driven and quantitative methods, an integrative physiology approach emphasizes concepts through experiments and observation across multiple scales [Gol19]. The multiscale nature of physiology allows us to, for example, link how signaling events at a molecular level lead to the normal, i.e., healthy, contraction of cardiac muscle in a normal heartbeat. An understanding of the normal processes and functions of the body allows us to recognize those that are abnormal, such as in atrial fibrillation, a heart problem where the upper chambers of the heart do not follow a regular beating pattern. With recent advances in hardware and software, as well as in experimental and imaging modalities, it is now possible to model many of these processes across several scales. Consequently, it is time for a discussion of visualization tasks and techniques for multiscale physiology. This survey provides a broad overview of common approaches and highlights research opportunities in visualization for physiology across multiple scales.
Modern clinical workflows involve a battery of tests and imaging protocols related to physiology. These are used to guide therapy, monitor disease progression or treatment response, and identify new biomarkers for medical research. Improved technology and hardware capture an unprecedented volume and diversity of data through models and simulations, e.g., advanced numerical simulations of blood flow, as well as through various acquisition techniques, e.g., fluorescence lifetime imaging microscopy (FLIM). Data range from 2D to 3D images, from static to time-dependent, from scalar to vector to tensor fields, and are often multivariate. The visualized physiological processes range spatially from nanometers to full body length and temporally from femto-/nanoseconds up to hours, months, and, in some cases, even years, as shown in Fig. 1. However, these data are often specific to a particular and relatively narrow spatio-temporal scale, and establishing links between these multimodal data types from the nano- to macroscale has been described as a grand challenge for many years from the perspective of systems biology [OGF*10b; ODo21], visualization [OYBH15; GSG*21], and in a multidisciplinary 2018 Dagstuhl Seminar [AGMN18]. Linking these data requires multi-disciplinary teams to develop analytical models and visualization approaches that can bridge the range of spatial and temporal scales. The Physiome/Virtual Physiological Human and affiliated subprojects [ABBC05; Hun06; FBC*08; TCA*11; VH16] have aimed to model processes that range from the molecular to organ scales, and beyond, to understand the multiscale interplay of physiology. The National Institutes of Health's Human BioMolecular Atlas Program (HuBMAP) aims to comprehensively map the human body at single-cell resolution from both a structural and functional perspective [Con19]. Numerous works have blossomed from these initiatives, such as OpenCMISS-Zinc [BBB*11], a library for building multiscale models and visualizations of physiological processes.
Despite the wealth of collected and simulated data for physiology, not all of this information can be, or is optimally, visualized through data-driven means. Hand-crafted medical illustrations are an alternative or supplement to data-driven visualization for representing physiology. Illustration remains ubiquitous when communicating information to a broad audience where simplification and abstraction of concepts are essential [GJ07; HG11; RMW14; SSHT14; JH14]. More generally, illustrations are invaluable in communicating abstract concepts, theories, and models. In this sense, illustration can provide a source of inspiration for abstraction in data-driven visualization. Paired with computer-supported solutions, an illustration can be brought to life through interactivity and adaptability to different scenarios. However, the time and labor cost for creating such illustrations prevents their use for, e.g., patient-specific data visualization. Throughout this report, we highlight select illustrative works to demonstrate opportunities where illustration can inspire or augment data-driven approaches.
Physiology has received extensive attention from the visualization community but in a fragmented, unevenly distributed form across subtopics, data sources, and visualization techniques. Few of these works extend their focus beyond one or two scales, e.g., only molecular [KKF*17], molecular and cellular [GOF20], or organ [LSBP18]. This paints a limited picture of the true multiscale nature of physiology. Similarly restricted in scope are surveys on a particular data type, e.g., PC-MRI by Köhler et al. [KBvP*17]. Technique surveys, such as by Bach et al. [BDA*17] on space-time cubes, McGee et al. [MGM*19] on multilayer network visualization, or Preim et al. [PM20] on medical animation, may span multiple scales, but physiology is often only one application area of many under discussion and is not the primary focus. Still others have explored the multiscale challenges in visualizing biomedical data from a high-level perspective [AMM*07; KH12; MMC*12; VRFW14; CJS*21], though without a specific focus on physiology. While our survey does not go into the level of detail that these surveys visit for their respective areas, the novelty of our work is in presenting a unified overview across multiple scales. We do so by discussing the coverage of these surveys alongside representative individual works that contribute to the same scale. We intend this report as an introductory resource for the space of challenges and opportunities for visualization research applied to physiology. Our framing for the work we survey additionally provides a different perspective than related work. We embed each article in a spatio-temporal context and draw from Brehmer & Munzner [BM13] to discuss its contribution according to the high-level user task(s) that it addresses.
Perhaps closest to our survey in terms of the scales of biological organization covered, Secrier & Schneider [SS14] discuss general visualization techniques from the bioinformatics domain for physiology from the molecular to population scale, but this review is brief and high-level. O'Donoghue et al. [OBC*18] review the use of omics and imaging data in biomedical research from molecule to population level for the primary purpose of exploration. However, their discussion is from a systems biology and bioinformatics perspective and mainly focuses on the visualization of molecular data to understand multiscale physiology. Our work covers a broader set of data types and a wider range of physiological processes.
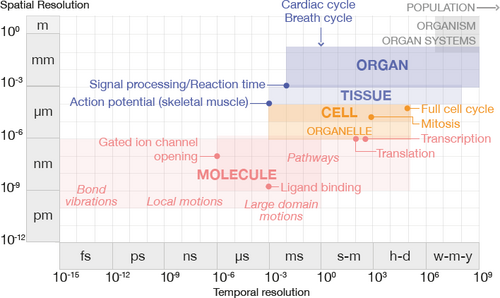
The general spatial and temporal ranges of human physiology, partitioned according to scale. Bold text indicates areas of focus in this survey, with example processes labeled in each scale.
To our knowledge, this work is the first of its kind to broadly overview the space of visualization for physiology that covers a scope similar to Lipşa et al.'s survey of visualization for the physical sciences domain (astronomy, chemistry, etc.) [LLC*12]. Our main contributions include:
- This is the first literature survey paper of its kind that provides a view into mature and open opportunities in visualization research for physiology. Our work surveys both within and beyond the core visualization venues.
- We focus the content of our survey on physiology topics that are highly-researched and cited both within the visualization community and in related physiology domains.
- We introduce a novel taxonomy that addresses these different topic areas and their respective opportunities by embedding works within a spatio-temporal context according to the high-level visualization task that they address.
In the following, we provide a brief background on physiology in Sec. 2, followed by a discussion of our survey methodology (Sec. 3) and classification structure (Sec. 4). Sections 5–8 are each dedicated to a spatio-temporal scale in our taxonomy, in order of increasing biological complexity. In each section, we first introduce the necessary background information and relevance of the physiological processes discussed at the given scale. We then provide an overview of visualization conventions and trends that we observe in the related literature according to task. A brief discussion of the mature and open challenges in visualization concludes each scale before transitioning to the next section. Fig. 2 provides an overview of our organizational approach for these sections. Readers interested in a specific topic, e.g., molecular pathway visualization for exploration, may easily navigate to the part of the report that is most relevant to their interests and needs. Section 9 provides an overview of true multiscale visualizations uncovered in our search and leads into a discussion of the challenges and research outlook on visualization opportunities for physiology (Sec. 10–12).
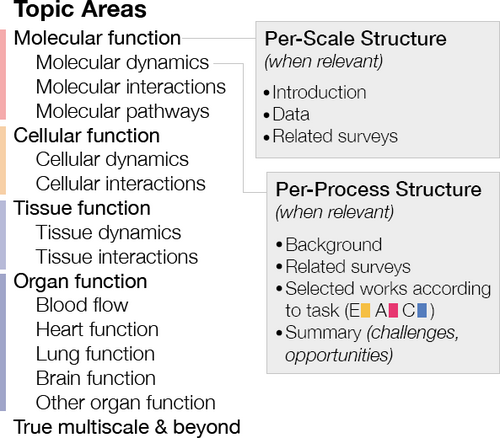
Our approach for Sections 5–8 organizes physiology topic areas in order of increasing spatio-temporal scale, with detailed per-scale and per-process points for each topic area (when relevant). Task categorization: E = exploration, A = analysis, and C = communication.
2. Physiology Background
Normal human physiology requires a careful balancing act, known as homeostasis, of numerous processes that occur over a broad span of time and space, as shown in Fig. 1. The smallest entity in the human body with the functional characteristics to sustain life independently is the cell. The cell itself contains molecules, such as water and ions, and organic molecules, such as proteins, that participate in processes necessary for its survival. Genes are the basic unit of heredity in cells that are made up of DNA and which encode the synthesis of RNA, which directs protein synthesis. Genes, proteins, and other molecules interact in sequences of reactions and interactions that are described as pathways. These pathways form networks and contribute to specific cellular functions. Molecular structures fall generally in the range of nanometers, and molecule-scale processes over a broad temporal range from femtoseconds, e.g., bond vibration between atoms in a molecule, to seconds, e.g., global motions or reaction sequences in a molecular pathway [HK07], to minutes and hours in the case of pathways involved in metabolism, gene expression, and signal transduction [ASP*18].
Human and other eukaryotic cells contain specialized cellular structures called organelles that participate in and facilitate the molecular pathways that keep the cell functioning. The mitochondrion is one type of organelle known as the energy powerhouse of the cell, while the nucleus is another organelle that provides the housing for our genes. A cell has the ability to communicate and exchange nutrients with its environment through its semipermeable membrane. This membrane contains specialized molecules, known as receptors, as well as channels and other structures that facilitate communication and exchange. Cell-scale processes relate to cells that average in the range of tens of microns in size, and with a temporal range of milliseconds, e.g., action potential generation, to minutes, e.g., mitosis, to a day for a complete cell cycle in humans.
Human cells differentiate into specialized cells with shared properties that group together to form tissue, which is classified into four different types: muscle, epithelial, connective, and nerve tissue. The physiological properties of these different tissues reflect the function that they serve. Cardiac muscle tissue, for instance, is responsible for the periodic contraction of the heart. Skeletal muscle tissue moves our limbs, while smooth muscle tissue moves food through the digestive system. Abnormal tissues occur where the comprising cells take on different characteristics than in normal tissue. For example, cancerous tumors have completely different tissue features than the surrounding tissue in which they occur. Tissue-scale processes span hundreds of microns to millimeters and temporally range from milliseconds, e.g., signal propagation, to weeks or even months, in the case of tissue growth and development.
Organs are composed of different types of tissue and perform major physiological processes according to their location, form, and composition. For example, one of the functions of the heart is to pump blood that contains life-giving oxygen and nutrients to the body's cells. Organ-scale processes that we consider in this report tend to fall within a narrow spatio-temporal window: organs like the heart measure in the range of centimeters, and the temporal range of a complete heartbeat or a complete breath cycle is in the range of seconds. Organs with similar functions are grouped into systems. For example, the cardiovascular system consists of the heart and blood vessels that are responsible, among other tasks, for carrying oxygen and nutrients to the body's cells. Our organ systems are interdependent. The cardiovascular system cannot function without the respiratory system, which includes the lungs, because the lungs handle blood re-oxygenation. The healthy functioning of an organism is dependent upon the systems of the body working in concert.
3. Scope and Methodology
This survey sketches out trends and opportunities in visualization for physiology across multiple scales, with an emphasis on human physiology. Fig. 3 provides an overview of our methodology.
Thematic Topics in Physiology. We restrict our survey to timely, highly-cited thematic areas in human physiology to ensure that our survey presents a relevant research agenda. For this, we used Web of Science's “hot papers” and “highly cited” filters with the keyword “physiology.” A “hot paper” is any paper published in the past two years that has received enough citations to rank in the top 0.1% of papers in its field. A “highly cited” paper ranks in the top 1% of cited papers for its field and publication year. To get a sense of the diversity of topics, we took the top 20 papers from each of these filters and excluded works that did not relate to humans or other mammals. We keyed these papers to topical area of physiology, e.g., molecular pathways or heart function, following standard medical physiology textbooks [HG11]. For a complete list of these papers and their topical areas, we refer the reader to Tables 1 and 2 in the supplementary materials.
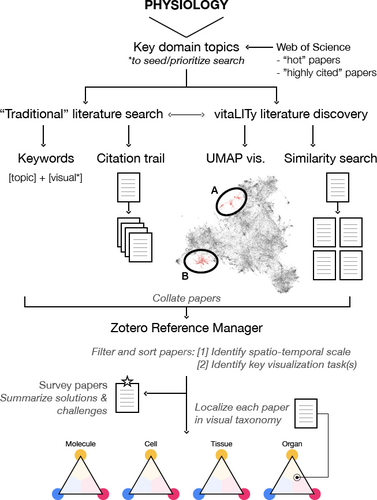
Our literature search process included both traditional search methodologies and vitaLITy [NKWW21]. VitaLITy's UMAP visualization allowed us to identify two main groupings of physiology-related visualization literature: (A) contains molecular-scale visualization literature, while (B) contains cell-, tissue-, and organ-scale works.
Search Criteria. Our survey focuses on visualization research for understanding physiology. We excluded pure method papers, meaning that the visualization literature we included must have a clear discussion of the domain science as a possible application for the proposed method. We also excluded works where the main visualization goal is to understand structure, although we included limited examples of instances where a physiological process is used to visualize a structure, e.g., 4D PC-MRI data to describe vessel boundaries [BKG*16]. We focused primarily on input data that is either itself dynamic, or is being used to capture snapshots of a dynamic process. We excluded purely longitudinal studies. We limited our search and discussion of research in areas that have already been well-covered in visualization and looked more comprehensively in less well-covered areas. In summary, we included application-oriented papers that center around a key topic area we identified from timely and highly-cited physiology research and that apply visualization in a novel way for the topic domain.
We focused our literature search on core visualization publication venues: IEEE TVCG, CGF, C&G, BioVis, VIZBI, and VCBM. The domain sciences may adapt visualization techniques in a novel way to interpret their data. We did not extensively review work published in domain-specific venues, but included a selection of works relevant to our included physiology subtopics to show visualization's use from this perspective. These domain-specific papers contributed to approximately 46% of the total literature collected.
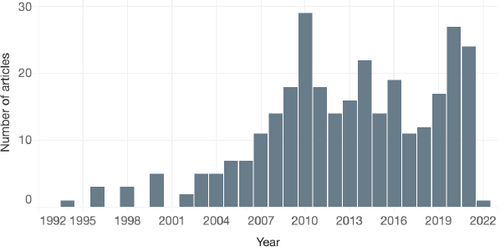
Distribution of papers (excluding surveys) according to publication year.
Search Process. We conducted our initial search using a combination of Google Scholar, PubMed, and IEEE Xplore based on keyword search [physiology topic] AND visual*. The literature search was divided between two coauthors.
We used vitaLITy [NKWW21] to complement our search, a recent visual analysis tool that allows for serendipitous discovery of academic literature. The vitaLITy database at the time of this writing consists of 59,000 literature items from 38 computer science venues that include our core venues listed above. These are searchable in a standard table that includes paper title, abstract, keywords, and authors, as well as a similarity search and a 2D UMAP visualization of the embedding space for the entire collection. For details on these tool features, we refer to Narechania et al. [NKWW21]. In the UMAP visualization, we identified two main groupings of literature, shown in Fig. 3, that helped focus our search: (A) groups works for visualizing molecule-scale processes: molecular dynamics, interactions, and pathways, while (B) includes works for visualizing cell-, tissue-, and organ-scale processes. Within each of these groupings, we searched for existing surveys and state-of-the-art reports to identify saturated topics. For example, since a number of reports have been written on visualizing different topics at the molecular scale, we devote less space to discussing this scale in our work and focus more comprehensively on scales and physiology topics with less coverage. UMAP exploration also helped us to identify relevant individual papers. We used works found in vitaLITy to seed our more traditional search approach and vice versa. This allowed us to perform a more complete literature search that accounted for terminology differences between domains.
Refining Process. In a second detailed pass of our collected works, we reviewed titles, abstracts, and figures to determine topical fit for our survey. At this stage, we used the publication year as a secondary check for our search coverage. If necessary, we revisited vitaLITy for topic areas that had a publication year gap and resampled papers from this time frame.
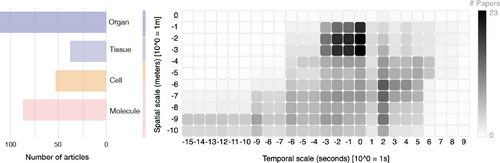
Distribution of literature by spatio-temporal scale, excluding surveys. Left: Literature are sorted by molecule, cell, tissue, and organ scales. This chart counts literature only once, according to the scale to which they contribute most. Right: Works are visualized in terms of density over spatio-temporal space, encoded by darkness. The horizontal axis shows temporal scale in units 10nseconds, while the vertical axis describes spatial scale in units 10mmeters. The dark region at the upper center indicates an abundance of works to visualize organ-scale physiology over the range of seconds, e.g., one heartbeat. The dark region at the right corresponds to works visualizing gene expression data.
Collection Summary. Our complete literature set includes 366 works, 61 of which survey or provide an outlook on an aspect of physiological data visualization. Approximately ⅓ of these works have been published in the last five years, with ⅔ of the total set published in the last ten years. Fig. 4 shows the distribution of works by publication year. The peak in publications in 2010 is a point we discuss in Sec. 10. Following literature collection, we classified all papers according to a two-level taxonomy to help identify challenges and opportunities in this domain. Due to limitations in space, we discuss a subset of these works in this paper, with the full library available in supplementary material and at https://lauragarrison87.github.io/star.web/vis_tool.
4. Taxonomy and Overview
Physiology spans the basic sciences and medicine, requires diverse domain knowledge, uses myriad data types, and employs a wide range of visualization techniques. Classification by domain, e.g., biology, chemistry, physics, or medicine, may seem the most obvious approach. However, these sciences are tied into each process and are difficult to classify separately, especially at the molecular scale. Molecular reactions are dictated by biology, chemistry, and physics and are core to disease diagnosis in medicine. In addition, different domains often adopt slightly different terminologies and classification systems. For example, biology distinguishes between gene, protein, tissue, organ, system, and body, while neuroscience follows a neurochemical, neuronal, region/network, and brain classification scale. This creates more confusion when organizing literature according to domain.
Although classification by data source may feel most natural in the context of the visualization pipeline, this does not provide a unified axis for the scales we survey. Simulations and models may span the molecule to organ scale but tend to be heavily focused on particular topics, e.g., heart function [ANL*16] or lung function [KBV*15]. While new imaging technologies, such as hierarchical phase-contrast tomography that maps organ to cell level, have come closer to realizing this possibility [WTW*21], no unifying technology yet bridges from the molecule up to the organ scale from a general physiological perspective.
4.1. Spatio-Temporal Organization
To minimize semantic collisions or confusion, we classify literature into scales along a spatio-temporal axis that is roughly discretized according to biological complexity: molecule, cell, tissue, and organ. This discretization is inspired by the organization of physiology textbooks [HG11]. Fig. 5 (left) shows the distribution of non-survey papers we collected that are categorized according to this scale. Works that span multiple scales are counted once for each scale, e.g., a work that we classify as both molecule and cell scale is counted in both the molecule and cell groupings.
We bundle temporality into this scale discretization based on the fact that, as structures increase in physical size, they tend to be involved in more biologically-complex processes that take more time to complete. This relationship between increased structural size/complexity and time has been discussed elsewhere in different domain contexts [SPW*08; SS14; DMRM17; GM17]. We can observe this phenomenon in Fig. 5 (right), which represents collected works classified in a range according to the scale that the input data spatially and temporally resolve to and up to the spatial and temporal scale of the structure and process of interest, e.g., the whole brain. For example, while EEG measures neural activity, we cannot visualize individual neurons with this modality, and that is not the intent of conducting these types of studies. Visualizations of EEG data fall in the organ scale. The relationship between space and time is not perfectly linear, as reflected by the dark groupings in the upper center and right regions of the chart. The upper part corresponds to organ-scale processes that occur over the range of seconds, such as a heartbeat or a full breath cycle, while the right part corresponds to the time for the expression of a single gene [MJM*10].
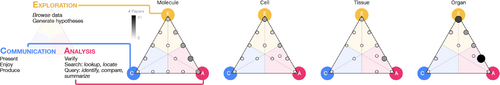
Left: Distribution of literature according to scale and high-level task, the latter of which is adapted from Brehmer & Munzner [BM13]. Right: Many visualization approaches support a combination of exploratory, analysis, and communication task(s). Darkness and size dually encode the number of works that are categorized with a given task combination within each triangle.
Our classification system does not formally extend beyond the organ scale for a few reasons. First, restricting the scales we examine keeps the scope of this survey manageable. In addition, our preliminary searches found limited visualization research that exists purely at the system– or organism–level, and the tasks and visualization techniques implemented are similar to those observed at the organ scale. We briefly review examples of works beyond the organ scale, as well as selected works that use true multiscale approaches, where the visualization aims to facilitate a task at three or more scales, in Sec. 9.
Although an enormous range of physiological processes occur at all scales of the body, we focus our survey on a few categories of processes at each scale that are timely for the physiology domain. Processes occurring at the molecule scale include molecular dynamics, e.g., the motion of atoms and molecules, reactions between molecules where electrons and/or atoms are exchanged, and molecular pathways, which describe a chain of molecular reactions. Cell-scale functions that we highlight include cell dynamics and interactions, such as how the cell develops and communicates with its environment. We include the dynamics of the cell's organelles at this scale, such as mitochondrial activity. Tissue-scale functions consider the behavior of aggregates of cells of the same type and include tissue dynamics, such as growth, and tissue interactions, such as signal propagation in neural tissue. At the organ scale we consider processes related to blood flow as well as the functioning of the heart, brain, and lungs. The body of visualization literature at this scale is large in correspondence to the maturity of data acquisition techniques available and the ease at which these processes may be captured or simulated.
Non-human studies present an issue in this measurement-based classification system. For example, a visualization of the neural pathways in a fruit fly brain concerns micrometers, while a human brain measures in centimeters. Although the focus of this survey is on human physiology, there is immense value in considering model organism physiology. These experiments tend to be more innovative, with correspondingly greater likelihood of exciting visualization opportunities. In cases where we include model organism physiology visualizations, we map the organism's scale to the human scale. Following this logic, we classify, e.g., a visualization of fruit fly brain activity at the organ scale.
4.2. High-Level Visualization Task
A subsequent layer categorizes the literature according to three high-level visual tasks: exploration, analysis, and communication, as illustrated in Fig. 6. These tasks are drawn from Brehmer & Munzner's typology of abstract visualization tasks [BM13]. We chose high-level, rather than low-level, tasks to provide a clear picture of the broad needs and challenges users face in visualizing physiology and how this compares across scales. We first considered categorizing works according to visualization technique, e.g., direct visualization. However, since task ultimately drives the chosen visualization technique, we feel that this is a more meaningful classification mechanism that furthermore has been the basis of classification in other surveys.
Exploration tasks often arise when the user is unsure of what the data contain. In the context of the data visualization pipeline, the user typically wishes to minimally abstract the data and produce a visual mapping that is as close as possible to reality. They do this to explore what the data actually contain. This is often a preliminary step in a larger analytical process. Analysis tasks occur when the user may be more sure of the intrinsic characteristics of the data, but now want to extract meaning from these data. Analysis often relates closely with exploration, where a user may begin with an exploratory approach to generate a hypothesis, then perform low-level analytical tasks alongside statistical methods to follow up on their hypothesis. In the visualization pipeline, analysis involves production of new artefacts through data transformation, derivation, and abstraction [AMM*07]. Although important for any task, audience is a key part of a communication task, where a visualization is created to underline key concepts of the data for presentation, education, or enjoyment to a particular group, whether to peers or to a broader audience. Visualizations developed for this task are often further abstracted from the data than in analysis- or exploration-oriented tasks, and can incorporate cinematic or storytelling elements to convey the author's interpretation of the data. While nearly all publications include figures to communicate scientific results, for this survey, we identify uses of visualization for communication beyond what is achievable with standard, out-of-the-box tools.
Many visualizations cannot be defined through strictly one of these tasks and rather are often generated to meet a combination of tasks. A work created to explore the data may also specify a visual analysis task. For example, ZigCell3D [dHKMK13] visually explores simulations of cellular functions while also providing tools for the visual analysis of the underlying simulation. The same data may be visualized for a communication task if the data are more visually abstracted or if annotations or glyph overlays are added to tell a story about the underlying information. This may also summarize key findings from, e.g., a visual analysis session for a broader audience. We apply weighted categorizations to each work, excluding surveys that cover many works. This produces a vector of three values between 0 and 1 for exploration, analysis, and communication, respectively. We then use this vector to position a work in a barycentric coordinate space, which allows us to compare and contrast between similar works within and between different spatio-temporal scales.
We arrange works graphically within a triangle where each of the three points corresponds to a high-level task, as shown in the left of Fig. 6. Exploration resides at the top of the triangle to reflect that, when exploring data, we are in a position of knowing the least about what we are looking for and/or the data are in their least abstracted form. Moving clockwise to the right corner is analysis, where we usually know something about the data and what we are looking for. Communication resides at the left corner, where data are highly abstracted and summarized in order to present, communicate, or serve data for enjoyment. The set of four triangular glyphs in Fig. 6 summarizes our visual taxonomy. Each triangle represents a single scale space, where the three triangle points represent the three respective visualization tasks. Circles indicate the position of each work as encoded by its balance of exploration, analysis, and communication tasks. Circle darkness and size dually encode the number of works with a given task categorization that we collected in our survey.
Literature Overview. The scale and task categorizations for each literature item collected for our survey can be browsed in the References section. Scale is labeled with a grayscale color tile, and related surveys are labeled with a black tile. Individual categorized works include a miniature bar graph that indicates the task(s) addressed, i.e., exploration (yellow), analysis (magenta), and communication (blue), from a range of 0 to 1. An interactive overview of the complete literature collection is available at https://lauragarrison87.github.io/star.web/vis_tool.
5. Molecular Function
All physiological processes depend on events that occur at the molecular scale. Molecules are the smallest units of a chemical compound and are themselves made up of atoms. Molecules in living organisms are known as biomolecules. Large biomolecules, known as macromolecules, include DNA, RNA, proteins, and lipids, while small biomolecules include metabolites [KKF*17]. Molecules are dynamic, flexible structures that interact and react with nearby molecules or ions. These individual reactions link into pathways with cascading effects at larger spatio-temporal scales.
Data. A number of data types can be used to characterize molecular function. Omics data, which is an umbrella term that includes genomics, proteomics, metabolomics, and transcriptomics data, are used experimentally to characterize and quantify molecular patterns and behaviors that scale up to the behaviors of cells, tissues, and entire organisms [SS07; GOB*10; GVS*20]. Some of these data may be utilized as structural data sources. These include nuclear magnetic resonance (NMR), x-ray crystallography, cryo-electron microscopy [KKF*17], and mass spectroscopy [MB19]. High resolution microscopy techniques, such as fluorescence lifetime imaging microscopy (FLIM), may also be used to visualize dynamic signaling events between proteins and their specific locations in living cells [SP15]. Molecular dynamics simulations commonly pair with these structural data to describe conformational changes and reactions between molecules [SML*10].
Related Surveys. Various aspects of molecular function have received considerable attention from the visualization community with a strong focus on visual exploration and analysis tasks. The main challenge with these large, multifaceted datasets is to balance insight with complexity. The BioVisExplorer tool by Kerren et al. [KKLS17] is a useful starting point to explore the space of methods for molecular data visualization according to data type, data properties, and data tasks. Alharbi et al. [AAM*17] contribute a brief survey of surveys of molecular visualization of computational biology data, where the main focus of many of the surveys included is on either structural aspects of molecules, or on visualizing molecular dynamics and interactions from simulations and structural data [OGF*10a; HGB14; KKF*17]. Visual analysis tasks for molecular interactions related to molecular cavity structure and dynamics have received considerable attention [KKL*16; SLD*17]. More recent surveys that include discussions of methods to visualize molecular dynamics and interactions of structural data include those by Schatz et al. [SKPE19] and Martinez et al. [MKA*19]. Johnson & Hertig [JH14] provide a communication-oriented guide for the visualization of molecular structural data with a short discussion on visualizing molecular dynamics.
Surveys centered around visualizing processes from omics data from a systems biology and bioinformatics perspective similarly emphasize the challenges of balancing insight with complexity for visualizing these data. The primary focus is often on data analysis, with exploration secondary [SS07; SBH*08; GOB*10; PMP*15; SSHU15]. These works provide an overview of data types, visualization tools, and methods for large-scale omics data. Their focus is on tools and methods for molecular interactions and pathways with the goal of understanding and interpretation, generally by experts. Approaches for using multilayer network graphs to visualize omics data are explored in McGee et al. [MGM*19]. From the pharmacology domain, Csermely et al. provide a comprehensive review of analytical tools for molecular interactions, pathways, and networks for the purpose of drug discovery [CKK*13]. Visualization approaches highlighted are limited to node-link diagrams with abstract glyph representations of molecular entities.
A number of works target the visualization of genomic data, where understanding patterns of gene expression is an important facet [NCD*10]. Nusrat et al. [NHG19] survey the tasks, techniques, and challenges for visualizing genomics data, of which gene expression and interactions are an aspect. They emphasize the need for tools that allow for exploration for hypothesis generation and follow-up analysis. Works by Goodstadt & Marti-Renom [GM17], Yardimici et al. [YN17], and Ing-Simmons & Vaquerizas [IV19] highlight several visualization methods that incorporate the 3D nature of gene organization in chromatin and chromosomes into the visual analysis of gene interactions and expression, which is of particular interest to experts in recent years.
In the following subsections, we review a selection of visualizations for three categories of processes that themselves increase in temporal and spatial scale: molecular dynamics, molecular interactions, and molecular pathways.
5.1. Molecular Dynamics
Molecules are flexible and dynamic structures that frequently transition between conformational states. These structural dynamics are due to interactions between a molecule's atoms, with nearby atoms from their environment, and environmental conditions like temperature and pressure [Rin18]. Molecular dynamics are characterized by the time scale of their conformation fluctuations (kinetics) and the amplitude and directionality of the fluctuations (structure). These fluctuations form a multidimensional energy landscape. Local fluctuations typically occur over nanoseconds, while global fluctuations can span microseconds to seconds. These global fluctuations are big conformational changes that signify protein-protein interactions, or reactions that initiate a molecular pathway, e.g., signal transduction [HK07]. Domain researchers are particularly interested in this energy landscape as it applies to understanding mechanisms of disease and for drug design.
Visualization approaches that target the flexibility of molecular structures often use nonphotorealistic visualization techniques that show molecular surfaces at atomic resolution. Ball-and-stick and ribbon visualization representations are also commonly used [JH14; KKF*17]. Color is often assigned to highlight differently-flexible regions. In addition, many visual analysis methods incorporate simple graphical elements, such as glyphs or path-line visualization techniques.
Approaches that are mainly exploratory in nature are intended to allow researchers to browse and familiarize themselves with the results of a molecular dynamics simulation. These approaches tend to use minimal abstraction and encodings that are familiar to the domain [GBC*14; LBPH10]. This also includes tools like VMD [HDS96] or PyMol [Sch15], which are widely adopted in the application domain. Visual elements may be used to draw out features within the data for exploration, such as pathlines to indicate atomic paths that drive changes in overall molecular shape [DRSR15].
As researchers become more familiar with the data, they may switch from exploration in an overview to analysis of a particular region of a molecule or molecular complex. Visual abstraction methods that exploit the hierarchical structure of molecules are useful to facilitate toggling between exploration and more focused identification and comparison tasks [LVRH07].
Analytical approaches tend to incorporate interactive techniques and/or statistical methods. These allow researchers to identify and compare specific information about how parts of the molecule are moving in relation to one another. These approaches are of particular interest for researchers in drug development and protein engineering. A structural visualization of the molecule is usually important alongside 2D plots showing, e.g., trajectories [GBB*19]. For example, Fioravante et al. [FSTR13] use principle component analysis to cluster molecules that have correlated motions, while Schmidt et al. [SBH02] derive mean shape conformations from the data to allow researchers to identify and compare metastable conformations. Other methods incorporate additional visualizations, such as time curve plots and heatmaps, to help researchers identify particular shape changes or constraints of interest [DCS12; TTL*15]. Interactive filtering techniques also help researchers identify particular movements of interest [HV00].
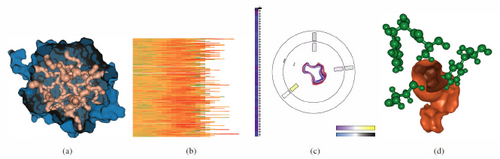
MoleCollar [BJG*15] enables the visual analysis of protein tunnel dynamics and biochemistry in large ensembles of molecular dynamics simulations. Reproduced with permission.
Conformation changes of a molecule affect not only its outer shape but also the shape of cavities or tunnels in the molecule. The shape of these tunnels affects the ability of a ligand, i.e., a signaling molecule, to travel to its binding site within a molecular cavity or tunnel [HG11]. These approaches usually include a mix of direct 3D visualization methods alongside heavily abstracted methods to accommodate a specific goal, e.g., to understand how the shape of a tunnel changes over time. Visual analysis methods include aggregating a molecular dynamics time sequence to a single contour plot [BLG*15]. Heatmaps to show variation in tunnel properties, such as tunnel centerline length, amino acid composition, and bottleneck size, can also be paired with direct visualization of a molecule [BJG*15; GHX*20], as shown in Fig. 7. More extensive visual abstraction from the original molecule shape can be used to understand dynamic structural changes and energy landscapes without occlusion [KBP*16; KFS*16; LAQS20].
Limited visualization research is dedicated to the communication of molecular dynamics, as much of this work comes from collaborations with domain experts with specific exploratory or analytical goals. Communication-oriented works use graphical elements, such as arrow glyphs, to illustrate molecular flexibility [BJG12]. Tools geared towards medical illustrators, such as Molecular Maya (mMaya) [Cla22], allow artists to animate molecular motions.
Summary. Most of the works visualizing molecular dynamics are targeted at domain experts for a combination of exploratory and analytical tasks. The time scale over which molecules change shape ranges over at least nine orders of magnitude. This presents a yet–unsolved visualization challenge to provide exploratory and analytical tools to experts to review and identify movements of interest in a vast temporal space.
5.2. Molecular Interactions
Molecular interactions can lead to an (ir)reversible reaction between two molecules [Ede13]. This can change the properties of the input molecule(s), synthesize a new molecule, e.g., polymerization, or destroy a molecule. Enzymes speed up the rate of a specific chemical reaction within a cell. Ligands form a complex with another molecule, often a protein, at a binding site [HG11]. This binding initiates a series of reactions. The time scale of molecular interactions is large, ranging from nano- to seconds, which presents a similar visualization challenge as we discussed in Sec. 5.1.
Similar to molecular dynamics, visual approaches to molecular interactions are strongly spatial and tend to focus on exploratory and analytical tasks for domain experts. Experts are often interested in exploring a simulation of interactions between molecules and in analyzing those interactions, e.g., protein-ligand interactions, that could lead to binding events that trigger a molecular reaction. Multi-view visualization approaches are ubiquitous, where at least one view typically uses surface models and nonphotorealistic rendering techniques to visualize the molecule(s) of interest at atomic resolution. Coloring of the molecule(s) is often according to biochemical properties or measures of uncertainty. Standard information visualization techniques, e.g., line, bar, and scatter plots accompany the spatial view to describe interaction energies and other important simulation parameters.
Key research questions relate to positional relationships between the protein and ligand, which influence the likelihood of binding. In some instances, the researcher wishes to observe such interactions in living cells, as Kerppola's work demonstrates [Ker06]. Detailed position and interaction information from structural and simulation data can be shown on a per-atom basis through direct visualization of structural and simulation data. Glyphs and color-coding on molecular isosurfaces often enrich the visualization with additional information [TWK*11; TMJ*15].
Works that target the identification of important interaction and binding events incorporate multiple data sources, e.g., simulation and mass spectrometry [MB19], often in interactive views with some level of guidance. Aggregation of trajectory data to aid the analysis process is common. In addition, navigational techniques help experts locate features of interest in these often large and highly complex datasets. Such techniques can allow users to reveal different levels of detail on-demand [AKCL19], incorporate focus+context techniques, as in the CLISD view of protein-ligand interactions by Schatz et al. [SFS*21], allow filtering of subsets of trajectories [JFB*19], and allow users to jump to different parts of the simulation timeline [DHR*18]. Visual abstraction of the hierarchical structure of molecules may also be exploited for the analysis of different configurations of protein complexes [FJK*19]. Many works visualize 3D molecular structures alongside interaction energies and other important molecular parameters. These parameters may be represented by glyphs as by Hermosilla et al. [HEG*17], or using scatter and box plots as by Furmanova et al. [FJB*16].
Works that emphasize the broad scale of space and time over which such interaction events can occur use adjustable aggregation measures to manage spatial and temporal complexity [BTM*19; PBBH19; VS21]. Other works eschew 3D structural information entirely in favor of abstracted graphics to visualize pairwise interactions of interest [VHG*18; ZLW*21; SC21].
Tools and techniques from medical illustration and animation can be used to explore and share possible hypotheses in modeling environments, such as AutoDesk Maya [NLHI20], or to communicate molecular interactions between experts or to other stakeholders. Approaches that give users tools to create molecular interactions through rule-based frameworks enable exploration and sharing of the resulting simulation data [GCK*18; NSK*21]. Guided, interactive exploration through a rule-based simulation to track interactions in a molecular environment allows users to see the direct output of the simulation results or understand the spatial context of reaction events between molecular structures. Some of these methods employ illustrative techniques, such as focus+context, that are approachable for education and outreach [MPSV14], and incorporate multiple temporal scales into the visualization [KPV*14]. Visual complexity of molecular interactions scenes is an ongoing challenge, but research has shown that oversimplifying the crowded environments in which such interactions take place can be counterproductive to learning [JM12].
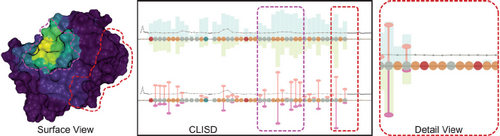
Figure 8 The Compressed Ligand Interaction Sequence Diagram (CLISD) [SFS*21] provides an overview of protein-ligand interactions over the course of a simulation, with unimportant residues visually de-emphasized and important residues and related parameter values given greater visual emphasis through size and color cues. Reproduced under Creative Commons CC BY license.
Summary. Exploring and analyzing molecular interactions is valuable for experts to understand and identify features and behaviors that can be used for pharmacological research. The main challenge to visualization is to continue researching effective methods that allow experts to understand the massive simulation datasets that are generated. This can be achieved via interactive tools to enable the identification of reaction events that occur very briefly within a temporal space that spans several orders of magnitude.
5.3. Molecular Pathways
Reactions between molecules create small changes in their immediate environment that trigger other reactions. This chain of reactions describes a molecular pathway [HG11]. Metabolism, signal transmission, and gene regulation and expression pathways are essential to life. Metabolic pathways describe the sequence of chemical reactions that occur in our bodies, such as the process for a cell to break down food into energy, or a pathway that builds a new molecule. Signal transduction pathways move a signal from the exterior to the interior of a cell with the help of proteins embedded in the cell surface known as receptors. Gene regulatory pathways turn genes on or off. When a gene is turned on, this allows the process of gene expression to occur, which transcribes and translates DNA instructions to create, e.g., a specific protein [Ins20]. Pathways do not exist in isolation and interact together in larger networks.
Selected visualization approches from Gosling, a grammar-based toolkit for scalable and interactive genomics data visualization [LWLG21]. Reproduced with author permission.
Understanding the participants, sequence, and timing of molecular pathways is key to understanding physiology at larger scales. Given the complexity of the input data, visual methods to address these goals tend to target expert user exploration and analysis tasks. These incorporate varying degrees of abstraction and interactivity. 2D information visualization techniques dominate, with networks being the most common technique, to show a sequence of steps in a pathway. Heatmaps, line plots, chord diagrams, and histograms are common for the visualization of gene expression.
The most straightforward visual methods allow experts to explore and identify the sequence of actors that participate in a given pathway(s) use node-link diagrams. Perhaps one of the most well-known pathway exploration tools, Cytoscape [SMO*03], uses node-link diagrams to visualize complex pathways and networks for users to explore and query. Such diagrams show entities in highly abstracted glyphs, often indicate reaction direction, and can indicate the location where the pathway takes place [LHM*09; KS20]. Brushing and linking [GVS*20], filtering [LPK*13], comparison [Sch03], and focus+context [JKL*10] techniques for detailed analysis are often supported. Numerous works have explored different layout algorithms to reduce crossover and clutter of these complex and often crowded visualizations [BALJ06]. Many implement graphical representations using, e.g., a subway map metaphor [LYKB08], that are approachable to broader audiences [CBF*15; KDE*15]. Visualizations of pathway simulations can be abstracted in 2D as line charts or heatmaps [SMW*21] to help experts to better understand the timing of pathways. An entirely different pathway simulation approach by Le Muzic et al. [MWPV15] employs an agent-based approach with 3D molecular structures to tell a multi-temporal scale story that provides insights to both experts and broader audiences alike.
Identifying and comparing levels of gene expression can provide valuable information to researchers on the activity of a given pathway, while studying gene co-expression can provide understanding of patterns and similarity of certain expression pathways. Gene expression and co-expression data are most commonly displayed in heatmaps, parallel coordinates, and chord diagrams, as shown in Fig. 9 [LWLG21]. Tools like Caleydo [LSKS10] enable the exploration and analysis of large-scale pathway data alongside gene expression data, using node-link diagrams and heatmaps in a 2.5D layout. OmicsTide [HFKN21] uses clustering with profile plots in a Sankey diagram to compare trends from gene expression and proteomic data. Some tools capture the multiscale nature of gene expression in visualizations that span the scale of individual nucelotides to entire chromosomes [MMP09]. Gene expression is a dynamic and fluctuating process. Other tools allow for exploration and analysis of temporal patterns of these fluctuations [MWS*10], and in some cases use clustering methods to facilitate pattern identification [CAM18].
Researchers are similarly interested in identifying and comparing concentrations of metabolites in specific locations of the body. This information provides another perspective on the activity of certain pathways. Tools for visual exploration and analysis of metabolite concentrations are useful to understand metabolic profiles of diseases at a molecular level [NLK*14]. Such approaches can use basic statistical methods alongside heatmaps [GVC*20], violin [JEG*19], or star charts [JML19].
Strong communication-oriented approaches to visualizing molecular pathways often draw inspiration from medical illustration and use cinematic elements to convey pathway information, such as Berry's animations showing the process of DNA transcription in real-time [Ber18]. In this way, the molecular dynamics and reactions between molecules at key steps in the pathway can be visualized in a larger context. Large charts showing pathway elements, when mainly used for communication, usually rely on abstraction of visual elements to create a scene that balances accuracy with readability [BVR*17; GMF*21].
Summary. Similar to molecular dynamics and interactions, the majority of visualization research works focus on expert-centered exploration and analysis tasks. The extraordinary complexity and volume of these data often necessitate guidance in interactive methods, and many approaches use statistical methods to reduce the analysis space alongside minimalist graphical elements. Further research into methods that facilitate a greater degree of exploration for hypothesis generation of these data, while managing the volume of information present, is an ongoing visualization challenge and opportunity for all molecular processes. Visual communication research for pathways is also important to develop further. Giving the public better tools to understand how diseases work, such as in COVID-19, can improve adherence and trust in public health protocols. Understanding physiology at this scale is essential, as molecular dynamics, interactions, and pathways work in concert to trigger behavioral and physical responses that form the foundation of cell physiology.
6. Cellular Function
The cell is the structural and functional unit of life in humans and many other organisms. Cells are self-contained, bounded by an outer membrane holding several substructures (organelles) that perform specific functions and facilitate molecular pathways that keep the cell alive and within balance [TBS*15]. We acknowledge that the distinction between cell-scale processes and molecular-scale pathways can be blurry, particularly in the case of large-scale molecular networks that themselves define cell physiology. We categorized each work according to the scale that is most relevant to the user's interest. In cases where interest is primarily in understanding whole-cell behaviors, we categorized the work in this scale, while if user interest is primarily in the various molecules that form a pathway or network, we categorized corresponding works in the molecular section.
Data. Input data to visualize cellular function can be acquired experimentally, often through different time-lapse optical microscopy methods on living cells and most commonly through fluorescence microscopy. This technique allows researchers to tag cells with specific proteins that fluoresce under the microscope, enabling visualization of specific cellular structures and behaviors. For a complete overview of live cell microscopy methods, we refer to Jensen et al. [Jen13]. Electron microscopy, which kills the cell, is often used to supplement live microscopy methods to visualize ultra-structure details inside the cell [Gla20]. Biomechanical methods to experimentally determine the effects of different forces on cells and their organelles include atomic force microscopy and tweezing [BGG*18; HFS21]. Omics data, e.g., single-cell RNA sequencing (scRNA-seq) data, which provides the molecular expression profiles of live individual cells, can also supply detailed information on cell function and behavior [NTM*21]. Because a cell is a self-sufficient entity, it is often a natural starting point for physiological models of cell behavior [SPW*08]. The CellML repository, maintained by the Human Physiome Project, is a rich repository for cell behavioral models [LLHN08]. Stochastic simulations are also useful to simulate complex biological pathways and networks within the crowded and dynamic environment of a cell and its surroundings [VTGB16].
Related Surveys. Surveys covering the visualization of cell dynamics and interactions are sparse relative to the molecular scale. Pretorius et al. [PKE17] identify six classes of visualization techniques: spatial embedding, space-time cubes, temporal plots, aggregate plots, dimension reduction, and lineage diagrams in their survey of visualization for live cell imaging. These techniques remain common in our report at this scale. Goodsell et al. [GOF20] provide a review of visualization methods that combine experimental data from microscopy, structural biology, and bioinformatics to build structural models of entire cells, mainly through nonphotorealistic visualization techniques. These models include details of molecular behaviors and interactions that contribute to cell dynamics. Feig & Sugita [FS13; FS19] review models for visualizing whole-cell dynamics at the resolution of the myriad molecular interactions that occur within a cellular environment. Their work highlights the use of surface, ribbon, and ball-and-stick molecular models at atomic resolution.
In the following, we discuss visualization trends and challenges for cellular dynamics, which essentially are processes that affect the cell itself, and cellular interactions, which are processes that involve a cell interacting with its neighbors.
6.1. Cellular Dynamics
The dynamics of a cell are dictated by molecular pathways and by behaviors of its organelles, which are themselves modulated by molecular pathways. These pathways drive the dynamics of a cell's organelles, the ability of a cell to move in its environment, the suite of internal mechanisms that dictate a cell's growth, and that lead to cell division and death, to name a few processes. We also discuss visualizations for whole cell models.
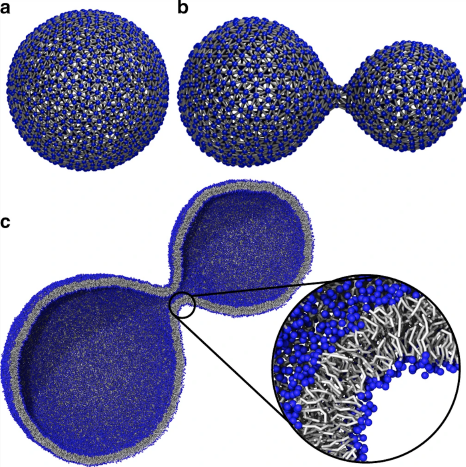
Visualization of vesicle formation from molecular dynamic simulation data [PKWM20]. Reproduced under Creative Commons CC BY license.
Organelles participate in and facilitate the network of pathways that drive the overall behavior of a cell. Visualization tasks related to organelles are often exploratory in nature, e.g., to observe the effects of an experimental condition under microscopy. Visualization methods from the domain often show time-lapse imaging data unmodified or surface renderings. This can help users to understand the shape changes a cell nucleus undergoes in response to experimental conditions [SKZ*15], the movement of cellular vesicles [RVT*20], or the compaction of chromatin in the nucleus over different phases of the cell cycle [OPD*17]. Analysis-oriented approaches often color-code regions of interest on volume-rendered segmented data or raw data slices to identify and compare features that indicate the functioning of underlying pathways [JLC*17; Gla20]. Glyphs are used to annotate features of interest on imaging data as a process occurs [WAM*20], and heatmaps can quantify interactions between organelles over the course of an experiment [VCL*17].
Simulations using 3D surface models can help answer questions about how organelle structures move and behave. For example, Waltemate et al. [WSB14] visualize membrane dynamics at molecular resolution in small “patches” of the cell membrane. More recent works visualize microtubule dynamics [KVGM19] or dynamics of mitochondria and cell transport vesicles [PKWM20], the latter of which is shown in Fig. 10. Such visualizations are adaptable for use in education environments, with systems like LifeBrush designed to explore mesoscale environments, e.g., the mitochondrial membrane, at molecular resolution in VR [DSJ19]. Even further toward communication are hand-crafted animations, such as the ground-breaking Inner Life of the Cell [Bol06], which shows the interplay between various organelles and molecules within the cell using cinematic techniques and visual abstraction to focus the narrative.
The individual dynamics and interactions of organelles influence and facilitate the cell's response to input from its environment and internal mechanisms that push the cell through its life cycle of growth, division, and death. Visualization of cell movements in 2D or 3D to discover and understand behaviors under experimental conditions is common in the domain, e.g., to understand cell cycle progression [SKM*08; HRP*12].
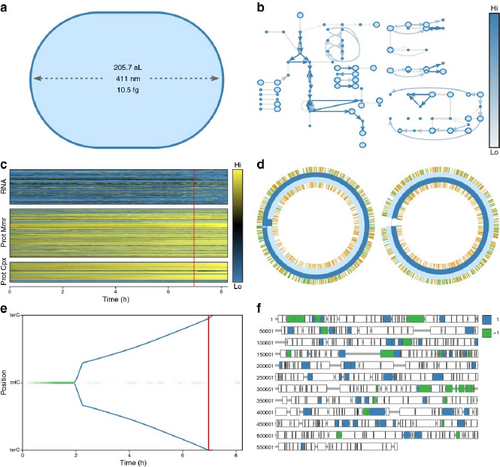
WholeCellViz [LKC13] modeling framework to explore and analyze simulations of cellular dynamics, such as cell shape, metabolic fluxes, and protein expression over the cell cycle. Reproduced under Creative Commons CC BY license.
Researchers may be interested in localizing subcellular structures and interactions through direct observation of imaging data [ASTM07] or may supplement imaging output with histograms, time plots, and similar aggregate visualizations to quantify features of interest [XMM*20]. Approaches may also use such aggregate visualizations to visualize results of classifications based on live microscopy alongside gene and protein expression data [BHK*21]. Dimensionality reduction methods, such as tSNE and HSNE, are useful methods to characterize and compare different cell behaviors and types from high-volume experimental data. Visualizations can map the results of these methods to a scatter plot, with color coding the cell type according to an expression marker [HPvU*16], and include guidance methods to more easily explore and analyze the data [HPvU*17]. Others deal with these high-volume data by identifying exemplar cells, either automatically or through user specification, to track specific derived attributes, such as growth, in response to drug treatments for cancer research in a multi-view visualization [LPJ*22]. Simulations allow researchers to manipulate cellular parameters for exploration and analysis. Simulations of parts of the cell cycle can be queried and explored for hypothesis generation in tools like CellCycle-Browser [BYG*18], which uses an interactive multi-view visualization containing heatmaps, line, and scatter plots to show the results of parameter changes to the system.
Whole-cell physiology is naturally multiscale, with limited works addressing visualization and specific user tasks at both molecular and cellular scales. These works enable experts to better understand intracellular functional and structural relationships. Highly abstracted approaches, similar to node-link network diagrams to represent molecular pathways, can visualize the multiscale interactions that occur within the cell [QHW*21] for querying or exploration. Visualizations of whole-cell simulations in 3D are useful to put molecular pathways into context, such as the effects of signal transduction on the cell's function [FKRE09; FKRE10] or the conditions and events that lead to cell death [FDSE11; FKE13; SBD*14]. WholeCellViz [LKC13] and ZigCell3D [dHKMK13], the former which is shown in Fig. 11, are whole-cell modeling frameworks. These frameworks allow researchers to explore and analyze cellular simulations in a biological context, from the molecular to the entire cell scale. They include pathway information as maps, as well as animation. ZigCell3D also incorporates imaging data and 3D models. A recent structural model of a whole Mycoplasma cell [MAK*22] provides an unprecedented means for researchers and the general public to explore and understand the structural and functional relationships of entities within the cell.
Summary. The visualization of cellular dynamics puts molecular pathway information into a cellular context and enables understanding of overall cell behavior. Experts are often interested in exploring and quantifying these data directly from imaging methods, with analysis of key features in aggregated plots. Further research into more interactive methods to facilitate analysis that allow experts to move away from simple rendering of microscopy data is a possible direction to explore. Very few works, especially from the visualization community, support expert study of organelle dynamics and behavior. This is an open space for visualization research. Multiscale visualization becomes truly meaningful at this scale to connect molecules with cellular behaviors. While numerous methods allow for exploration of whole-cell physiology, analysis of such models remains relatively limited, and this is another future research opportunity. Finally, in some contexts, communication-oriented approaches can serve both experts and a broader audience equally, as cells are less conceptually-abstract entities than molecules. Research into such approaches, particularly with regards to public health and in facilitating conversations on the mechanisms of disease, is an exciting challenge.
6.2. Cellular Interactions
In reality, cells do not exist in isolation. Their physiology is strongly influenced by interactions with their environment and neighboring cells. In this section, we discuss works that focus on the behavior and fate of individual cells, where understanding the environment and neighboring cell interactions are key to the user task.
As in previous topics, many visualization works are born out of collaborations with domain experts and mainly address exploratory and analytical tasks. These methods allow researchers to browse experimental, imaging, or simulation data to understand cell communication, lineages, and migratory patterns.
Direct visualization of live cell imaging data provides an overview of cell division, adhesion, signaling, and movement patterns within different environments, e.g., tumor microenvironments [EEA*08]. Clustering methods can facilitate user exploration of intercellular communication networks from single-cell transcriptional data. COMUNET classifies and clusters cell types according to ligand-receptor pairs and visualizes these communication patterns in node-link diagrams [SS20]. Migration patterns of differentiating cells can be understood by visualizing, e.g., spatial transcriptomics data, as in sci-Space [SRB*21] (Fig. 12), where heatmaps of gene expression patterns are measured over pseudo-time to capture cell differentiation and migration in a tissue context. Clustering methods can classify cells according to their migratory and other behaviors from microscopy data, which is valuable for comparative analysis [FHWL12]. Simulations with simplified 3D spherical models to represent individual cells provide information and control on per-cell properties of division, adhesion, and other environmental variables in vitro [Pal08; GHF*18].
sci-Space measures patterns of gene expression over time to understand patterns of cell differentiation and migration in neural tissue [SRB*21] Reproduced with permission.
Cell lineages contain valuable information on patterns of cell division, growth, differentiation, and death over generations of cells. This is particularly important with stem cells, which have unique regenerative abilities that have massive implications in cancer and other areas of medical research. Statistical methods to make sense of these patterns, in combination with the branching tree structures of lineage diagrams, help researchers identify and compare factors that influence cellular genealogies [GLHR09; PKE15]. These approaches can include visualization of cells within a spatial context, with navigational tools to observe how cells divide and where they migrate, e.g., to neighboring blood vessels [WWB*14; SGAT21]. Uncertainty due to segmentation of microscopy data when tracking cell aggregation is a challenge. Tools like Uncertainty Footprint [WH17] attempt to visualize and quantify these uncertainties for domain experts.
A growing interest in public science education has led to the development of tools like Bioty [WSGR17], a real-time programming environment that visualizes cell interactions for non-experts. As for other topics, hand-crafted medical illustrations are used to educate audiences on cellular interaction processes, such as the communication between a neuron and muscle cell [Goo09].
Summary. Visualizing cellular interactions adds a degree of complexity to cellular function visualizations, as these leave the self-contained environment of the cell to include external parameters that increase the complexity of the system. Collaborations with experts provide a means to explore and analyze data acquired either experimentally or through simulation, where gaining an understanding of the data through exploration is equally important to more targeted analysis tasks. Developing methods to facilitate exploration and analysis of cellular microscopy and lineage information through visual abstraction while retaining expert trust is one research opportunity. We found few works visualizing cellular migration and adhesion, particularly from within the visualization community. Given the importance of these behaviors in normal development and disease, this is yet another research opportunity.
7. Tissue Function
At the tissue scale, we see groups of cells of the same type that perform a specific function. These form tissue, which allows for coordinated behaviors to accomplish tasks impossible for single cells to perform. A tissue region also includes a container, known as an extracellular matrix, that holds the cells together and provides structural stability [SPW*08]. Each of the four main tissue types in the human body serves a specific role: (1) epithelial: covers and lines body surface and cavities; (2) connective: protects and supports body structures, i.e., organs; (3) muscle: coordinates movement; and (4) nervous: facilitates communication of nerve cells through electrical signaling. The visualization works we discuss in this section aid user tasks where the goal is to understand the overall behavior or dynamics of cells in aggregate, rather than individual cells. One process that we highlight includes tissue growth, also known as morphogenesis. This process drives the development of, e.g., blood vessels or tumors. We also discuss methods for visualizing perfusion, the delivery of nutrients to tissue via small blood vessels called capillaries, and the propagation of electrical signals through tissues.
Data. Spatially-resolved gene expression data can characterize the overarching physiology and behavior of tissue [NTM*21]. These methods may pair with imaging methods, such as seq-FISH+ [WNMR20]. For a comprehensive discussion of specific experimental methodologies, we refer to Waylen et al. [WNMR20]. Imaging methods for perfusion are well-established. These include positron emission tomography (PET), single-photon emission computed tomography (SPECT), computed tomography (CT), Doppler ultrasound, dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI), dynamic susceptibility contrast MRI (DSC-MRI), phase-contrast MRI (PC-MRI), ASL (arterial spin labeling), and optical methods such as widefield or fluorescence microscopy. Conventional widefield microscopy is common for visualizing tissue histology, although this requires fixing cells to a slide that not only kills the cells but can damage their spatial organization. Simulations and models often describe signal propagation within nervous or cardiac muscle tissue. Imaging data at the resolution to visualize individual neurons involved in signal propagation typically come from microscopy. State-of-the-art techniques for imaging brain tissue include confocal laser point-scanning microscopy (CLSM) and spectral precision distance microscopy (SPDM) for their high resolution, improved signal-to-noise ratio, and removal of out-of-field fluorescence. For further reading on these techniques, we refer to Tröger et al. [THP*20].
Related Surveys. Visualization works that cover tissue-scale physiology are limited and generally motivated by the needs of experts in the medical domain. Preim et al. [POM*09] survey methods for the visual exploration and analysis of perfusion data. The authors highlight cine-movies, subtraction images, and color-coded parameter maps on a single slice as basic visualization techniques. Advanced visualization techniques covered include multiparameter visualizations, e.g., colored height fields, combining structural information with dynamic perfusion data, or extracted features, e.g., temporal curves. Schlachter et al. [SRM*19] survey visual computing methods for radiotherapy planning, which include a detailed visual analysis of the metabolic profiles of tumors that can be acquired through perfusion data. Volume visualization techniques that fuse multiple data sources into a single image through overlays and color-coding are common in this area, particularly within the application domain. More advanced visualization techniques enable exploration and analysis of uncertainties in segmentation, or analysis of perfusion parameters in parallel coordinates, scatter, and star plots, among others. Qutub et al. [QMK*09] review modeling efforts for angiogenesis from an application domain perspective, some of which include molecular and cellular-level processes in the resulting visualization that we discuss further in Sec. 9. Visualization techniques used to illustrate these models include node-link diagrams, line plots, and histograms to describe and analyze model parameters. Spatial visualizations include algorithmically-generated surface models and stacked image slices. Color maps to model parameters, e.g., tissue oxygenation or upregulation of a particular pathway.
In the following, we provide an overview of visualization approaches and challenges for tissue dynamics, or the behavior of aggregates of cells in a given tissue type. We then discuss tissue interactions, including the delivery of nutrients and the passage of electrical signals through specialized tissue.
7.1. Tissue Dynamics
Tissue dynamics refers to the behavior of aggregates of cells in a given tissue type. Visualizing tissue dynamics simulations allows researchers to explore the general mechanisms of tissue growth and development under changing environmental conditions. Understanding spatial relationships at this scale is particularly important, and many visualizations represent simulation data as 3D surface models to capture the development of blood vessels (angio-genesis) [QMK*09], liver tissue regeneration [HBB*10], embryonic limb [CHC*05] and organ development [DKY98; CGN*10], and in a non-human case, wing development [BWB*12]. Exploring changes in skin tissue as a response to aging in 3D is also of interest [IAJG15], with the ability to change parameters to simulate the impact of disease or dehydration. In addition to visualizing normal processes, simulations are valuable sources to visualize pathologies related to tumor growth under changing environmental influences [TMS*11; TRM*12].
Although we have discussed gene expression data previously for the analysis of cellular interactions, this family of methods is useful for tissue-level visual exploration and to identify biomarkers in cell aggregates when performed in situ and paired with imaging data. These cell aggregates are often identified through clustering methods [SIL*20], and can then provide molecular and cellular resolution maps of the body, e.g., of embryonic tissue development in the first trimester [BGC*17]. Subsequent exploration of such tissue maps provides an opportunity to discover emergent properties at this scale. Multeesum [MMDP10] exemplifies this interplay between exploration and analysis, where comparing similar expression profiles of aggregates of cells allows researchers to form hypotheses about gene relationships and location. Numerous domain approaches also use visualization mainly to confirm hypotheses, e.g., the direct visualization of digital histology slide data to quantify the progression of liver tissue damage in fibrosis [LLJ*21].
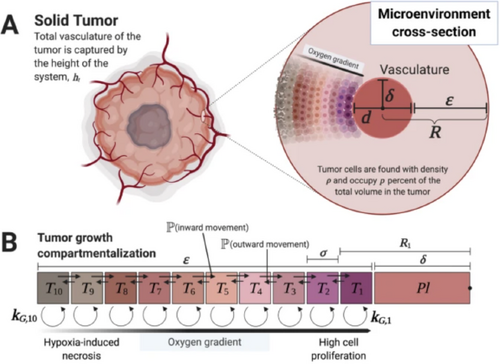
Compartment diagram and parameters for tumor growth [MMF20]. Reproduced under Creative Commons CC BY license.
Communication-oriented visualizations of tissue dynamics may take the form of adjustable simulations with easy-to-use interfaces and simple graphics that appeal to both researchers and a broader audience [WT21]. Animation of 3D models is also useful as an educational tool for showing the process of organ development, e.g., of the developing heart [SADC02]. Lastly, hand-crafted illustrations that describe models of tissue growth, as shown in Fig. 13, are invaluable to clearly and succinctly share models with peers [MMF20].
Summary. Visualization of tissue dynamics is often geared first towards exploration to familiarize oneself with the data, as data at this scale are typically complex and high-dimensional. Comparison tasks between groups are then common, where experts wish to identify parameters or biomarkers that define certain tissue behaviors or functions. Tissue dynamics are challenging to visualize in vivo, with approaches often using underlying processes such as gene expression or the presence of other biomarkers to characterize tissue functional properties. Simulations provide the means for visualizing truly dynamic growth processes in healthy and disease conditions. However, they often are abstracted from reality, with visualizations that expose only the final part of a multiscale story that is rooted in the molecular scale and with limited interactivity. Devising methods to enable fully interactive exploration and analysis of tissue dynamics, whether purely at the tissue scale or extending across scales, is a grand challenge and opportunity in visualization research.
Paraglyder is a tool for the visual analysis of tumor metabolic profiles [MHBS20]. Image provided by the authors and reproduced with permission.
7.2. Tissue Interactions
In this section, we focus on the interactions between different tissue types that allow for the passage and exchange of nutrients, as in tissue perfusion, or for the passage of electrical signals, as in signal propagation.
The function of blood flow on the microscopic scale is to supply, or perfuse, tissues in the body with oxygen, nutrients, and hormones and to transport waste products away into the appropriate “recycling” centers such as the lungs, kidneys, and liver. Different tissues have different perfusion rates, and visualization can be a powerful tool in profiling tissues based on these data. Perfusion data are particularly useful in identifying the extent, composition, and metabolic profiles of tumors.
Most use cases to visualize tissue perfusion are highly clinically-motivated with a particular set of analysis questions already in mind, although many methods incorporate a degree of exploration. These approaches incorporate structural visualizations of tumors and the surrounding tissue to provide context, and use derived multi-parametric imaging data to classify and visualize key physiological parameters. Approaches can use simple color overlays with parameters mapped to color channels [ABBM05] to quickly quantify values. Advanced visualization approaches allow for user interaction and exploration that incorporate time intensity curves [OPH*09; GPTP10; MNM*16], radar plots [MHBS20] as shown in Fig. 14, and scatter plots with glyphs encoding further information [MWH*20]. These representations allow experts to identify and compare features of tumor physiology.
Agent-based simulation of large-scale pyramidal neuron cell growth with the BioDynaMo platform [BHdM*21]. Reproduced with author permission and under Creative Commons CC BY license.
In the human central nervous system, information is processed by signal transmission and propagation between neurons, where the extracellular space plays an important role in transmission and signal propagation.
In visualizing signal propagation, particularly in simulations, experts wish to understand the mechanism, path, and timing of these propagation events. Straightforward visualizations that plot spikes in signal propagation are relatively common in the domain literature, such as in Rhodes et al. [RPR*20]. Microscopy data often provide a structural foundation for visualizing simulations of signal propagation between neurons or in a multi-neuron network [LHH*12; BHdM*21]. We show an example of a multineuron simulation network from BioDynaMo in Fig. 15 that is realized through procedurally-generated surface models. Dimensionality reduction methods can facilitate exploratory visual analysis of signal data, and allow users to identify patterns that signify, e.g., key points of a behavioral task [BKSP11]. Abstracted 2D plots, such as L-plots proposed by Dunin-Barkowski et al. [DLO*10], allow experts to observe and compare neural signaling patterns. Signal propagation is influenced by several factors, e.g., the distribution and density of glycogen around a synapse, which is the space where two neurons meet. Abstractocyte [MAB*17] is mainly designed for the visual analysis of astrocyte structure and distribution around neurons, but its pipeline includes glycogen distribution analysis.
Simulations of signal propagation are not limited to neural tissue. Visualizing simulations of the electrical conduction system in cardiac muscle tissue is of interest for experts to understand the timing and rate of signal propagation in different phases of the cardiac cycle [HBC06; BCG*11].
Summary. Unlike perfusion of tissue, the majority of work we found for spatially visualizing signal propagation comes from simulation data. As hardware and software continue to advance and support more complex simulations, there will be a corresponding increasing need to provide visual methods to explore, analyze, and communicate these data to various stakeholders. In tissue perfusion, visualization research that further supports exploratory analysis to identify complex biomarkers is an ongoing challenge. Visualization approaches for both tissue perfusion and signal transduction tend to have strong analytical components, especially in the case of perfusion, where clinical diagnostic improvements are the driving need for these applications. Communication-oriented research works are limited at this scale. The clinical motivation for understanding physiology becomes even more apparent at the organ scale, which deals with the interplay between different tissue types on a larger scale.
8. Organ Function
The last scale that we review is that of visualization for organ-scale processes. Organs are discrete units in the body that perform a function or a group of functions [SPW*08]. In the following, we cover visualization for four areas that are well-known in medical visualization: the dynamic properties of blood flow (hemodynamics), and the functioning of the heart, lungs, and brain. We also include a brief discussion of other visualized organ functions to give a sense of further opportunities at this scale, e.g., skeletal muscle function.
Data. Typical data inputs for visualization of physiological data at this resolution include a host of imaging modalities that can be time-resolved alongside, or separate to, simulation data. In many instances, structural data provide context to the visualized dynamic process. Information about anatomical structures can also be based on measures of physiology. For example, while diffusion-weighted and diffusion tensor imaging (DWI/DTI) methods use the diffusion of water molecules to capture the fiber architecture of the brain, these data are primarily used to probe white matter microstructure [PKB*14]. As such, the visualization of DWI/DTI data alone is out of the main scope of this work. The same is true for other such structurally-focused modalities. 4D computed tomography (CT) and a range of MRI sequences, e.g., dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI) and phase-contrast MRI (PC-MRI) [KBvP*17], are frequently used to assess various organ functions. Specialized ultrasound (US) methods are adapted to capture particular processes, e.g., Doppler ultrasound for hemodynamics, or electrocardiography [XT10] and echocardiography [MVM*11] for heart function. Computed tomography angiography is useful in assessing hemodynamics and heart function [MBK*10]. For a discussion of the strengths and weaknesses of these modalities in hemodynamics imaging, a hot topic in visualization research, we refer to Jennings et al. [JRG08], Markl et al. [MFK*12], and Sengupta et al. [SPK*12]. Typical imaging modalities measuring brain function include electroencephalography (EEG), functional magnetic resonance imaging (fMRI), and PET. We refer to Pfister et al. [PKB*14] for a detailed discussion of each of these modalities. Electromyography (EMG) [BHGG12] and motion capture data are common sources for assessing muscle function.
A number of these modalities alone have insufficient spatial and/or temporal resolution to capture an organ process of interest. These are often augmented with simulations, or simulations are developed from these imaging data. The purpose of simulation can also be to correct issues with the acquisition, such as motion-related artifacts. Approaches include computational fluid dynamics (CFD) for blood flow [RNNC15], statistical heart and lung motion models, and large-scale simulations of signal propagation for brain function.
Related Surveys. Surveys that discuss aspects of organ physiology on a broad level are typically motivated by the medical domain. Preim et al. [PM20] survey the use of medical animations for organ-level processes that tend to focus on communication-oriented tasks. Birkeland et al. [BŠH*14] survey works that fit in the ultrasound visualization pipeline, where the end-user task often is to explore the data and to identify specific features within the data related to, e.g., blood flow and heart function. Many visualization approaches at this scale combine modalities to overcome individual modality limitations. Lawonn et al. [LSBP18] provide an extensive discussion on multimodal visualization. Tory et al. [TRM*01] provide a brief overview of methods for MRI in combination with dynamic SPECT data. In general, visualization tasks at this scale focus on giving experts, whether in medical research or more directly in the clinic, tools to explore and analyze physiological features for improved diagnosis and treatment.
8.1. Blood Flow
While we previously looked at blood flow from the lens of how it supplies nutrients to tissues (Sec. 7.2), researchers often are interested in the dynamics of blood itself as it travels through the heart and vessels of the body. Understanding patterns of blood flow can help researchers and clinicians make better decisions about patient health, such as when to operate on an aneurysm.
Related Surveys. This is a mature area with several surveys and state-of-the-art reports available. For further details on visualization techniques and challenges on this topic, we refer to reports by Markl et al. [MKE11], van Pelt et al. [vPV13], Vilanova et al. [VPP*14], and Stankovic et al. [SAG*14]. For further reading on visualization methods specific to PC-MRI blood flow, see Köhler et al. [KBvP*17]. Most recently, Oeltze-Jafra et al. [OMN*19] survey trends and challenges in visualizing medical flow data, where the primary focus is on blood flow data. These surveys highlight a mix of exploratory and analytical visualization tasks, where tasks are highly motivated by domain experts' needs to locate and identify flow features that impact patient health. General flow visualization techniques are commonly used, e.g., glyphs, textures, integral curves, line integral convolution (LIC), colored cut planes, extraction to surface models, e.g., streamlines. Contextualization of blood flow dynamics using image slice or volume rendering of the surrounding anatomy is key in nearly all blood flow visualization scenarios [OMN*19]. Advanced visualization techniques often incorporate multiple interactive views with facilities for validation, filtering of key parameters, and uncertainty analysis.
Exploratory tasks generally aim at obtaining an overview of flow patterns and often precede a quantitative workflow. These may visualize simulation data [RNNC15; NCW09], use techniques that combine different modalities to create the visualization [PH07; BFS*09; FFI*12], or use single imaging methods [BdHdK*16]. The containing structure of interest, e.g., a vessel or the heart, is useful to preserve for context. The paths and direction of flow data can be presented as pathlines [dHJEV16], streamlines [BFW*11], arrows [HPT*08], and pathlets [ASN*14], which can also be used for analysis. Newer approaches have experimented with effects like smoke or dye to better visualize time-varying flow patterns with indications of uncertainty [dHLJ*19]. Interactive elements, such as virtual probes [vPBB*11], aid in expert exploration of flow patterns prior to quantitative analysis.
Although illustrative techniques are often associated with communication-oriented tasks, most illustrative techniques employed for this topic are aimed at experts to facilitate exploration of flow data. These approaches can, e.g., reduce occlusion from vessel walls through adaptive surface visualizations [GNKP10; LGP14] or focus+context flow lens treatment [GNBP11]. Other illustrative approaches facilitate exploration of wall thickness relative to flow properties [LGV*16].
Method of aorta straightening for occlusion-free comparison of flow over time [AH11]. Reproduced with author permission.
Many analytical approaches from the domain are limited in interactivity. These approaches again include a structural context, with similar visual representations for flow data as for general exploration. Visual representations also often use heatmap overlays and/or glyphs of the same styles mentioned for exploration, e.g., streamlines or arrows, to indicate flow velocities at particular points. This allows for feature quantification directly from imaging data [TSS*08; HPT*08]. Experts also often wish to quantify and compare flow rates in simulations relative to time-resolved imaging data [LGH*19].
Analytically-focused tools and methods developed from collaborations between domain experts and the visualization community often take more experimental or abstracted approaches to aid analysis tasks. For example, Angelelli et al. [AH11] flatten 3D tubular flow to 2D to compare flow patterns over time, as shown in Fig. 16. Semi-automatic classification and clustering methods are also common to aid expert identification and comparison of vortices and shapes in the data [OLK*14; ERH16; MBP*16; OCJP16]. Interactive linked views, particularly when including simulated and acquired data, can help experts to better evaluate hemodynamic patterns and model accuracy [LNHL20]. Multi-view visual analysis tools often rely on an interplay between exploration and analysis for users to get a sense of the data and browse interesting regions before identifying and comparing features to understand, e.g., aneurysm rupture risk [MWPL20; MVG*21].
Approaches from the research community to present blood flow in education for a more general audience are limited. These methods may hint at the original flow data through animation but instantiate red blood cells to indicate to a broader audience what the flow represents [GMF*21], or employ more fanciful metaphors to show the passage of blood in a cardiac cycle [CHV*14].
Summary. Despite the extensive work on hemodynamics, further challenges and opportunities remain. The ultimate aim of many of these works is to make visualization of hemodynamics available in a clinical setting to aid in rapid and accurate identification of life-threatening flow behaviors. Studies that assess the possibility of real adoption of these techniques in clinical routine are an interesting avenue to explore. Communicating these data then to patients, in a way that is both understandable and minimally-alarming, is essential and remains an open challenge in visualization.
8.2. Heart Function
Heart function is well-characterized in physiology and visualization research. In this section, we focus on visualization related to (1) the mechanics of the heart as a pump and (2) the cardiac conduction system of the heart, which is an electrical network that controls heart rate and rhythm [HG11]. Diseases related to these aspects of heart function include (1) myocardial ischemia, where the heart tissue does not receive enough blood from its supplying arteries, (2) heart failure, where the heart is unable to pump blood effectively, and (3) atrial fibrillation, a dysfunction of the cardiac conduction system that leads to irregular heart rate and rhythm. These diseases provide strong clinical motivation and drive many of the visualization use cases in this topic.
Related Surveys. Nazir et al. [NKA*19] survey the visualization of various aspects of heart function from the medical domain, focusing mainly on analysis for use clinical routine. Walton et al. [WBT*14] provide a broad overview of the methods and challenges in visualizing cardiovascular magnetic resonance imagery for clinical research before presenting a prototype approach for visualizing this type of data. Generation of surface models and volume renderings of the heart, paired with time-lapse video to describe deformation, are key visualization techniques for this topic. Heatmap visualizations commonly indicate parameters of interest, and the bull's eye plot is ubiquitous for the visual analysis of perfusion data to understand heart function.
Exploration-centered visualizations provide an overview to experts of general features and parameters related to shape changes, e.g., for specific chambers, valves, or the entire heart, in a cardiac cycle. These works visualize phases of the cardiac cycle from simulations on surface models (which typically are abstracted from acquired data), such as the LFX Virtual Cardiac Model [JJDS04] or the constrained Multi-linear Shape Model [IGV*10]. Patient-specific approaches visualize models in combination with acquired data [WWZS10; ANL*16], or only acquired data. Some works build predictive models, and preserve links between the simulation and the original data to understand the mapping procedure [SLS09]. Although we discuss blood flow extensively in Sec. 8.1, for completeness, we note that several approaches visualize patterns of blood flow to explore questions related to heart function, e.g., Kulp et al. [KMQ*11].
Approaches geared towards a combination of exploration and analysis, or focused more purely on analysis, often favor colormaps applied to mesh or imaging data to quantify parameters of interest. Rainbow colormaps are common, especially in the medical domain. These works often use multiple interactive views to link simulation, imaging, and derived statistical information. The main purpose of the visualization is to evaluate particular parameters, e.g., strain rate [HSTS98; PP00]. Evaluating the movement of particular landmarks can be aided by additional plots, such as parallel coordinates and heatmaps, to compare different motion parameters [THS*17], as shown in Fig. 17.
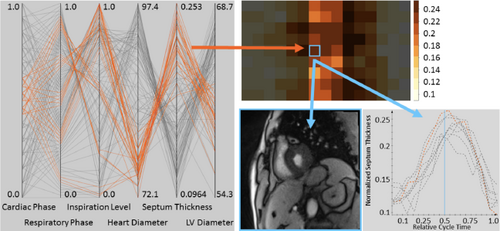
Visual analysis for the quantification of septum motion from real-time cardiac MRI [THS*17]. Reproduced with permission. © 2017 The Author(s). Eurographics Proceedings © 2017 The Eurographics Association.
Perfusion data, which we visited previously in Sec. 7.2, are often used as a basis to determine heart functionality. Most use cases are tied to experts with a need to identify and quantify particular tissue properties in the context of a pathology, and while exploration is a component, it is usually not the main task. Many approaches incorporate a bull's eye plot in cases related to myocardial ischemia, where heart tissue does not receive the blood and nutrients it needs to function. This visualization is familiar to clinicians to quantify the extent of the damage to the heart tissue, often alongside structural representations of the coronary arteries [OGHP06; TBB*07; TBB*08; RBPJ16]. The bull's eye plot can be further adapted for targeted visual analysis of the motion of the left ventricle of the heart over time [SCK*16]. Approaches are often interactive to provide an exploratory element for the user. Glyphs can be incorporated, as by Meyer-Spradow et al. [MSD*08], to quantify local tissue perfusion across the whole heart. Statistical, e.g., PCA, and aggregate measures may be used to reduce the complexity of the data [ODH*07] and include representations of uncertainty [RBPJ16].
Visualization is also used to evaluate the accuracy of simulations against acquired data, where heatmaps [MBK*10] or juxtaposed line plots [HOR07] indicate shape prediction accuracy. Other methods focus heavily on patient-centered care and outcomes [XSZ*16], such as simulations of surgical procedures, e.g., mitral valve clipping, with quantitative evaluation through heatmaps to help predict patient outcomes [MVM*11].
Some heart simulations have been developed, not only for expert exploration, but for use in education and surgical training. These include Dayan et al.‘s [DOS*04] 3D animation of the dynamics of a simulated mitral valve and the virtual reality (VR) simulation of radio frequency ablation by Pernod et al. [PSRD10]. Pernod et al.‘s approach uses a heatmap to show membrane propagation potential, a tissue-scale process, on the heart surface. VR has also been used for patients in a biofeedback scenario to manage stress [GWZE18].
Summary. While full cardiac models of heart function for detailed exploration and analysis are of high interest to the medical community, visualization research efforts to aid these tasks are relatively limited in comparison to the efforts dedicated to blood flow. In the analysis of perfusion data for whole-heart pathologies, a higher volume of works focus on solving tasks for clinicians. However, studies on their actual adoption and utility in a clinical setting are limited. Communication-focused works that visualize heart function, while numerous in the field of medical illustration, remain a comparatively limited topic in visualization. A number of recent works to visualize heart function rely on multiscale models. We discuss these works in Sec. 9.
8.3. Lung Function
The main function of breathing, i.e., respiration, is to provide oxygen to the tissues in the body and to remove carbon dioxide [HG11]. We focus here on the in- and outflow of air from the lungs and on limited cases where research includes other organs affected by lung movements.
Experts interested in learning about the features of lung deformation during breathing often rely on simulations, commonly from statistical modeling, to visualize this process [SG05; SID*08; KLO10; EWSH11; WWLS13]. These generally integrate with imaging data to provide spatial context, often via surface modeling or volume rendering techniques. More recent approaches have used neural networks to reconstruct lung deformations as surface meshes from 4D CT data [WZH19].
Experts are also interested in understanding patterns and features of dynamic airflow. This interest may be in visualizing airflow patterns within bronchial tubes [STM08] or on a larger scale. Kim et al. [KBV*15] present a coupled model of tissue deformation on the level of the whole lungs alongside network airflow, enabling predictions of various dynamic flow properties. The model is multiscale, but, as shown in Fig. 18, does not necessarily provide spatial visualization of cell-level air exchange and lacks visual interactivity. Wiechert et al. [WCRW11] couple tissue- and organ-scale processes to allow visual exploration of tissue regions locally and the whole lung and airway system globally.
In more analytically-focused cases, visualization enables the comparison of movements of an object of interest against an acquired signal [LCPS10]. Similar to heart function, visualization is also used to evaluate model accuracy against acquired data and to assess and compare the magnitude of lung deformation using heatmaps [NTC*19].
A coupled model of tissue deformation and network airflow that enables predictions of dynamic flow properties [KBV*15]. Reproduced with permission.
Respiratory visualization models, in addition to parameter exploration, may also be used to compare characteristics of air flow in healthy versus diseased patients [KBV*15]. In other instances, experts use lung function to indirectly provide information in the analysis of other organs. For example, breathing exerts force on the kidneys. The degree that the kidneys are compressed during the breath cycle can be used to evaluate kidney fibrosis [SHHB16].
Summary. The bulk of literature we found from visualization and domain research focuses heavily on expert exploration of airflow and lung deformation over breath cycles, with limited tools for analysis and even more limited work in visualizing lung function for communication. These works primarily come from outside the visualization community and represent an open opportunity to develop visual methods to support experts in better understanding and analyzing lung function. These tasks are particularly important with the advent of COVID-19, as experts work to understand the long-term effects of this disease on lung function.
8.4. Brain Function
The brain is part of the central nervous system that contains more than 100 billion neurons. It is the primary seat of control for any process occurring within our bodies. Understanding brain function provides a key to understanding human behavior as well as neurological diseases and disorders. While our discussion of signal propagation in Sec. 7 focused on the propagation of action potentials between cells, we now discuss signal propagation and functional neural connections over the entire brain. This is known as functional connectomics, where the brain is modeled as a network [STK05].
Related Surveys. Margulies et al. [MBWG13] and Pfister et al. [PKB*14] provide an overview of ways that the human connectome, both structural and functional, can be visualized for different exploratory, analytical, and communication tasks. Node-link diagrams, scatter plots, dendrograms, and heatmaps are common techniques in the application domain to visualize synchronous activity between brain regions. Structural models often provide spatial context for functional connectivity and can be depicted as image slices, surface models, volume renderings, or, in the case of DTI data, through advanced techniques that include ellipsoid or brushstroke glyphs [LAK*98] and superquadric glyphs [Kin04].
Experts are interested in learning how different regions of the brain functionally connect and in exploring patterns of brain activation in response to the presence, or absence, of certain stimuli. This represents a broader-scope view of the questions experts have when studying signal propagation. Heatmaps superimposed onto imaging data or derived surface models to show activation regions is a common approach that allows experts to explore and evaluate functional imaging data [FC00; WVE*03; RTF*06]. Interactive exploratory methods include dynamic querying for structural and functional connectivity using DTI and fMRI data [SAM*05].
Visual analysis methods help experts identify functional parameters and connections of interest. Standard methods often use a correlation matrix to identify functional connectivity [PKB*14]. Interactive analysis approaches often incorporate linked views that incorporate structural data alongside plots containing additional functional information, e.g., time plots [JNM*09; Lun10] or radar plots [MMH*13]. Color-coded isosurfaces from structural imaging, with network and scatter plots to show functional connectivity and correlations [DVF*10], are another example of interactive visual analysis interfaces. These interactive, multi-view approaches may facilitate hypothesis generation in addition to confirmative analysis and integrate several data types and plots [JBF*19]. Clustering methods can group structural fibers from DTI data into functionally-meaningful bundles. These can then be color-coded to aid identification and comparison of functional groups [GGZ*13], or to identify and compare resting state networks that are again color-coded [VMH08]. Other classification approaches to aid analysis use contrast subgraphs, shown in Fig. 19, as a primary means to compare between groups [LBG20].
Approach for classifying brain networks based on extracting contrast subgraphs, i.e., a set of vertices whose induced subgraphs are dense in one class of graphs and sparse in the other [LBG20]. Reproduced with author permission.
While the bulk of visualization research for brain function is targeted at experts, some works that use illustrative techniques can be used for communication to a broader audience. The work by Jainek et al. [JBB*08] exemplifies such an instance for their use of glows and a soft, harmonious color palette to show brain activity.
Summary. Visualizing brain function is challenging. This is owed to the high computational power needed to simulate activity over an entire brain, and the fact that many visualizations of brain activity are driven by imaging data that only indirectly indicate brain activity and function. An additional challenge is that functional networks and structural connections do not always overlap. Visualization research to depict these uncertainties can aid neuroscience researchers. Furthermore, tools tend to focus on tasks related to exploration and analysis for domain experts. The development of visual methods targeted toward clinical rather than research use to identify aberrant patterns of brain function is an ongoing challenge. Finally, as previously discussed, further research into communication-oriented approaches that facilitate doctor-patient communication and patient understanding are essential for raising the bar of health literacy and public health. For example, data-driven approaches can communicate public safety stories, such as the impact of alcohol on brain function when driving a vehicle.
8.5. Other Organ Function
Although we focus our survey mainly on blood flow and functions related to the heart, lungs, and brain, we briefly highlight other organ functions that are representative of further opportunities in visualization research for physiology.
Skeletal muscle plays a crucial role in body movement and defining anatomical shape. Lee et al. [LGK*12] review visual approaches, which often use 3D surface models, for modeling muscle deformation and simulation of skeletal muscle functions at this scale. Experts are chiefly interested in understanding the shape deformation that skeletal muscle undergoes during contraction and relaxation [BHGG12]. Visualizations represent muscles at different degrees of abstraction to serve different objectives in a simulation, e.g., a single action line to show the axis of movement [NT98] or reconstructing and simulating a subset of muscle fibers that capture the overall shape of the muscle as it deforms [KK14; KČ20; RMS20]. Visual analysis approaches are often interested in quantifying muscle properties during contraction, such as muscle stiffness [SSG*10] or muscle speed in contraction [ANB*03]. These data can be captured in multi-view visualizations that combine structural models with line plots that describe displacement, velocity, and acceleration over the course of the simulation [YGV*13].
The stomach and liver are part of the gastrointestinal system whose main functions are (1) to take in food and liquids and break them down into a usable form and (2) to remove waste from the body. The stomach is a highly elastic organ that serves as a temporary holding place for food and is responsible for its initial breakdown. To understand the stomach's changing dimensions over time, Gilja et al. [GHØB96] present a multi-view system that includes a planar map of a region of the stomach for experts to better understand these temporal changes. The liver is the centerpiece for many essential bodily functions, including blood and nutrient storage, with a complex physiology that could greatly benefit from visualization. Lin et al. [LJH04] visualize fluid transport through the liver and its vasculature at the organ and tissue scales, using simulations in combination with imaging and animation techniques. Their aim is to understand how the liver absorbs and metabolizes substances.
Metabolic activity, as discussed in the context of molecular pathways in Sec. 5.3, can be visualized on the organ level to assess for normal organ function. Approaches can be analytically-focused, as in Nguyen et al.'s [NEO*10] method to visualize uncertainty from PET kinetic modeling. Ropinski et al. [RVB*09] exemplify a more exploratory approach to visualizing organ-level metabolic activity. Their approach is also designed to facilitate doctor-doctor communication through interactive closeups, whereby users can adjust view layout and composition to best fit their communication agenda.
Summary. Organs function as the result of a chain of processes that begin at the molecular level and extend through the cell and tissue scales. While blood flow, heart, and brain function are especially well-covered in visualization research, the lungs and other organs that we briefly highlighted in this section have received less attention, although experts clearly benefit from visualization tools to aid their exploratory and analytical questions. As in other scales, we observe a comparative lack of visualization research oriented to communication tasks. This represents an open opportunity for future work. Furthermore, while visualizations of organ-scale processes are often highly clinically-motivated, relatively limited research investigates visual methods to aid practicing clinicians that are usable in a time-crunched environment.
9. True Multiscale and Beyond
Cakmak et al. [CJS*21] define multiscale visualizations as those that allow users to present, navigate and relate data across multiple abstraction scales. In this section, we highlight examples and trends of true multiscale visualization for physiology, where the spatial and temporal representations and associated user tasks span three or more scales within our taxonomy.
The Visible Human Project is one of the earliest multimodal data initiatives to visualize the human body in its entirety. Although the project mainly focuses on anatomical structures, one of its key goals is to link structural image data with text-based physiological data [Ack98]. Mathematical modeling initiatives to simulate multiscale human physiology like HumMod [HBH*11] are a rich resource for physiological models. The HumMod Browser, built using empirical data from peer-reviewed physiology literature, relies on grouped word clouds to allow experts to explore hierarchical and causal relationships of whole body physiology [WCPH13].
Works that are similarly exploratory, but include spatial information in their visualization, include Insley et al. [IGP11]. They present a multiscale, multiphysics simulation spanning from cell to organ scale of blood clot formation within a cerebral aneurysm. Their method allows the visualization of individual red blood cells, platelets, and solvent particles. It expands further to visualize large-scale flow patterns with streamlines, enables the observation of platelet aggregation along the aneurysm wall, and shows this phenomenon in the context of the surrounding vasculature. Miller et al. [MTT21] present a multiscale, although primarily structurally-focused, brain map that spans from molecule to tissue scale. They include functional information from spatial transcriptomics data to describe pathological Tau proteins as well as signal propagation information. Leggio et al. [LLC*19] present MorphoNet, an open-source online tool allowing users to interactively explore the anatomy and dynamics of biological entities from molecule to whole-organism scale. It furthermore allows for genetic data to be overlaid onto these models. Primarily focused on developmental processes, this tool uses 3D color-coded surface models and is targeted at research and education, as shown in Fig. 20.
Qutub et al. [QMK*09], whose work we briefly discussed in Sec. 7.1, present a review of multiscale modeling approaches, from molecule to organ scale, for angiogenesis. Although many of these models are multiscale, the visualization result is often not multiscale. An exception to this, although still limited and not interactive, includes Mac Gabhann et al.'s [MJP07] multiscale muscle model. This model includes muscle fibers (muscle cells), the microvascular bed that supplies oxygen to these cells and tissues, and the associated molecular pathways for angiogenesis.
While several truly multiscale models for the heart range from the level of ion channel opening to the heart's contractions over a full cardiac cycle, the visualization output often is limited to an organ-scale mesh representation. A heatmap then encodes electrical activity mapped to the surface mesh and static, non-interactive charts display the changes that occur at the cellular, molecular, or tissue scales [GRW*00; SDP*14; ANL*16]. Gil et al. [GAB*19] expand on this typical representation by incorporating myocardial fibers, extracted from DTI data, with simulation data to understand ventricular muscle tissue structure and connectivity. Chabiniok et al. [CWH*16] provide a review of multiscale cardiac modeling methods with an eye toward their integration for analysis in clinical practice. This shows a clear interest in understanding such models from the domain.
MorphoNet is an open-source online tool to explore and communicate the anatomy and dynamics of biological entities from imaging and genetic data [LLC*19]. Reproduced under Creative Commons CC BY license.
Some multiscale lung function models span from molecular to organ scale, such as those by Burrowes et al. [BDS*13], but lack integrated visualizations across all scales. As presented in Sec. 8.3, Kim et al. [KBV*15] propose a partially integrated visualization that captures the airflow dynamics of their model but lacks spatiality across levels. Furthermore, the visualization, as in many of these systems, does not allow for direct user interaction.
Multiscale models for brain function are becoming increasingly common as computational power increases. Spanning from cell to organ scale, such models capture the depolarization of a single neuron, signal propagation through brain tissue, and the effects that this signal has on brain function [ESC*12]. However, the visualizations of these models are often limited in the scales they depict. For example, in the Cognitive Computation Project, Ananthanaryanan et al. [AESM09] simulate a cat brain and visualize parts of the model with a 2D heatmap that plots groups of neurons with similarly-timed firing rates in a cortical area. A topographic plot provides a detailed view of the first signal spike within each neuron group.
Increasing works target the developmental stages of entire organisms through communication-oriented tasks that present physiology for education and outreach. Sorger et al. [SMK*16] use visual abstraction techniques to show, at a molecular level, the transitional stages of an HIV-virion. On a larger scale, the OpenWorm and its various subprojects [SLP*18] aim to visualize and simulate multiscale anatomy and physiology of C. elegans with numerous tools designed for public outreach. On a system-specific level, full-body virtual anatomical models of the human musculoskeletal system are a lively research topic, not only for domain experts [MLPE14] but for visual effects animators as well [RMS20]. Multiscale modeling approaches, such as those by Rzepecki et al. [RVV*14], propose to combine multimodal structural, physiological, and biomechanical data sources in an interactive viewer that scales from visualizations of cartilage tissue porosity up to a simulation of human gait.
10. Discussion
This survey is intended as a guide for visualization researchers interested in understanding common approaches and challenges to visualizing physiology from a spatio-temporal and task-oriented perspective. In this section, we discuss general themes along with lessons learned.
vitaLITy. Our literature collection approach utilized traditional search methods and leveraged new visual analysis tools from the community to facilitate this process [NKWW21]. Using vitaLITy was a huge help to identify holes in our search. Although vitaLITy does not span the space of literature that we covered, its coverage of the visualization literature is comprehensive, and helped us identify whether any holes in our search were due to issues with our methodology or due to the lack of visualization literature for a given topic. For example, using this combination of search methods, we discovered that the spike in publications in 2010 could partly be attributed to the rise in interest in the visualization of omics-related data, with four surveys published on the topic that year [GOB*10; NCD*10; OGF*10a; OGF*10b] along with the running of funded research projects like the Physiome, IllustraSound, and PhysioIllustration projects in Europe during this time frame. Close to 40% of the papers included in this survey were found using vitaLITy. In many cases, we used papers found with this tool as seed papers in a standard search methodology. Using this combination of tools helped give us confidence that our search methodology was even, in spite of an apparent bias towards, e.g., organ-scale functions. Through vitaLITy, we were able to sanity-check that these are heavily-weighted topics in visualization for physiology.
Spatio-Temporal Distribution. Classifying literature along a spatio-temporal axis uncovers a few interesting patterns in Fig. 5. Most salient is the dark grouping of works related to organ function that is positioned symmetrically on both spatial and temporal axes. The data acquisition methods to capture these processes (brain, heart, lung function, and blood flow) are well-established with strong clinical motivations. It is not surprising to see an abundance of work in this region, given these two factors.
Another, though less dark, region we observe ranges spatially from large molecules to cellular substructures and occurs over minutes. This corresponds to the abundance of work where the key process of interest in the visualization is understanding gene expression and where the majority of experimental methods, e.g., next-generation sequencing and proteomics methods, lack the temporal resolution to capture the active process of translation and transcription [DDM17]. These capture the result of the process, which occurs typically in the span of minutes (for a single gene) [MJM*10]. This density of work reflects the comparatively recent developments in experimental methods for measuring gene expression, which similarly accounts in large part for the increase in works over the last few years, as we observe in Fig. 4. We expect to see a continuing increase in such works.
The light region corresponding to a temporal resolution of a few seconds (101 sec) represents a few possibilities. One relates to the temporal resolution of experimental methods for gene expression: the majority of next-generation sequencing methods are, at best, on the scale of minutes [DDM17]. Another reason is that, while a number of processes bridge over this time range, for human physiology we found limited processes that are confined to this range. Diffusion of molecules across a cell can occur in this time span [MJM*10], but these are typically of interest when bundled into the greater context of a molecular pathway or in the dynamics of an entire cell, which encompass a larger time frame.
Smaller-scale processes, particularly at the molecular scale, have a much broader potential temporal range than the organ-scale processes our study examined. As many works in our survey allude to, this broad temporal range is both an enormous challenge and opportunity for visualization to aid experts in exploration and analysis of their data when events of interest are easily lost in temporal noise. A major and related ongoing challenge is to integrate into a visualization the larger spatial scales that build from, and are affected by, the molecular process of interest. The discussion of the differing breadth of time scales for the different spatial scales introduces another point: while temporality generally increases with biological complexity [SS14], living organisms are not bound to a system based on powers of ten, but rather to a roughly 24h cycle known as the circadian rhythm [MYS12]. This explains, in part, the temporal ranges of functions such as gene expression and the heartbeat and breath cycles that we observe in Fig. 5.
Cell & Tissue Function. We found comparatively few cell- and tissue-scale visualization works. This reflects the history and trends in the availability and technological advancements of the source data. Data that can truly visualize dynamic, living cell processes on the scale of molecule, cell, and tissue have only just recently become available and accessible, and we see in the publication dates that cell-related visualization works are on the rise. Computational power is also steadily increasing to the point where whole-cell visualizations are becoming a reality, while the additional complexity inherent in tissue-level physiology visualization is still a challenge. Furthermore, many application domain approaches use only basic visualization techniques, often simply reviewing microscopy imaging data, to explore or quantify features or behaviors of interest. This may show a lack of trust in abstraction that is, on some level, unavoidable when processing data to visualize through other methods. Approaches that incorporate raw imaging data in multi-view interactive tools alongside uncertainty quantification are useful directions to continue investigation.
Imaging and Simulation Data Across Scales. Visual exploratory and analytical tasks at the organ scale and, to a lesser degree, at the tissue and cell scales are typically closely tied to imaging data. Cases using a model at these scales usually compare or validate the model against imaging data. Hence, visualization tasks at larger scales are often to understand a given process from imaging data. This is not necessarily the case in real-time visualization of dynamic processes at the molecular scale, and in many instances at the cellular scale. Here, the visualization of a model often serves as the primary means to understand a given process. As technology improvements lead to higher resolution imaging methods, or if simulations come to be seen as more accurate and trustworthy than the imaging methods themselves, we may see this balance shift.
Task Distribution. Task distribution between exploration, analysis, and communication differs slightly across all scales, with one major trend consistent across all: communication-oriented visualization works for physiology are limited relative to exploration- or analysis-oriented works. Most visualization works we surveyed have concrete expert collaboration partners with specific data types and specific goals to understand these data. This works at a much lower level than communication. There are some possible explanations for this disparity in task distribution. One is that the data being visualized is often cutting-edge, and the domain scientists producing these data may not yet fully understand it themselves–visualization is needed to discover and analyze, and the communication element comes after understanding. A second possibility relates to data-related permissions. Patient and research subject data is often heavily protected and, in many instances, may not allow visualization beyond internal, domain-specific analytical use or require significant processing efforts to anonymize the data before use. Lastly, visual communication of this type of data simply is hard. The data are highly complex and multifaceted, and visual communication requires a high degree of understanding on both the side of the domain expert and the visualization researcher to distill this information into a clear narrative.
Application Domain Adoption. Many of the visualization approaches discussed in this report have not fully permeated the application domain. These are often highly-specific algorithms or techniques, while widely-adopted approaches in the domain are often more generally applicable. Furthermore, such production-ready, i.e., stable, solutions are continually maintained and designed for ease of use. For example, in visualizing molecular dynamics, tools such as VMD [HDS96] allow for easy visualization of simulations through a movie-like series of time steps. Across all scales, visualization methods adopted in the domain remain relatively restricted to the direct visualization of imaging data in a time-lapse sequence, as in light microscopy or medical imaging, e.g., fMRI. Techniques can extend into volumetric rendering with limited features for exploration and analysis, with the option to create surface meshes from these data. These visualization methods are easily available in tools like Amira [SWH*05], 3DSlicer, [FBK*12], and ParaView [AGL05]. Additionally, across all scales, basic visualizations such as bar or scatter plots are common to identify the frequency or distribution of physiological biomarkers under study. These are often created in tools like Microsoft Excel. Such charts are often limited in interactivity, e.g., only support filtering, and do not apply across multiple views. Coordinated efforts with funding agencies to establish initiatives for the deployment of advanced visualization techniques can enable broader access from the application domains. Furthermore, researchers can consider opportunities to develop advanced visualization approaches as plugins to existing domain tools rather than as stand-alone solutions. This is a possibility to enable greater domain accessibility, and can mitigate the resource limitations introduced by stand-alone tools.
11. Research Outlook
Physiology is a challenging, complex domain that visualization can do much more to contribute to in the coming years. Increasingly sophisticated modeling and data acquisition methods can capture physiology at finer spatial and temporal resolutions but, in exchange, produce even higher volumes of complex data. In addition, the multiscale, multisystem, multidisciplinary nature of physiology needs visualization to help bridge gaps, not only in exploration and analysis of data between scientists but in communication to a broader audience.
Numerous recent technological advances in imaging and experimental methods pose exciting opportunities for visualization across several scales. While light microscopy was previously limited to a maximum resolution of 200 nm, Pulsed Interleaved MINIFLUX with a standard microscope has increased this resolution to 1 nm, allowing visualization of metabolites and other small molecules in vivo [MSZ*20]. The boundaries of computed tomography have been similarly expanded with hierarchical phase-contrast tomography (HiP-CT) to allow true multiscale imaging from the organ down to the cellular level [WTW*21]. Other data have not received much attention from the visualization community, such as the suite of methods used to assess cell biomechanical forces [BGG*18]. Pioneering experimental methods to observe protein translation occurring in real-time in living cells, such as nascent chain tracking (NCT) [DDM17] are available, but visualization of these methods is limited. Expanding visualization research collaborations into such areas to develop new methods for experts to engage with these data is an enormous opportunity.
To answer questions left by gaps in systems biology and integrative physiology, research is shifting to focus on the human organism as a complete integrated network. This considers not only the scales that have been the primary focus of this report but also the coordinated efforts between organ systems and sub-systems. The study of the human organism as an integrated network is termed network physiology, with a set of grand challenges in this new discipline published only recently [Iva21]. Visualization methods and tools that can meet the exploratory, analytical, and communication demands for this area of study are exciting opportunities.
Multiscale computational models are increasingly ubiquitous with advancements in computational power and parallel processing. However, while these multiscale models exist, corresponding multiscale visualizations are often lacking or exist in unlinked, un-integrated forms. This is another opportunity for visualization research.
True multiscale and multisystem approaches necessitate multi-disciplinary collaborations. A dearth of visual methods and tools facilitate knowledge transfer between domains or communicate physiology to the public. As the last two years of the pandemic have demonstrated, clear and accurate visual communication of physiology for public health is critical at all levels of society.
12. Conclusion
This survey offers a broad overview of visualization trends and opportunities for physiology and aims to provide a foundation for discussion and future research directions in this area. From a mixed-methods literature search approach using state-of-the-art visual analysis tools, we embed our discussion of these approaches in a spatio-temporal context that focuses on the core tasks driving the visualization: exploration, analysis, and communication. Our report demonstrates an abundance of work at the organ scale, particularly for hemodynamics. Molecular visualization, particularly related to visual analysis and exploration of actors in molecular pathways, is a growing research area driven by the advent of new technologies. These new technologies hold immense promise for visualization research that incorporates multiple data types to span the true multiscale nature of human physiology, from molecule to organ scale and beyond.
13. Acknowledgements
This work was supported by the Visual Data Science for Large Scale Hypothesis Management in Imaging Biomarker Discovery (VIDI) funded by the University of Bergen and the Trond Mohn Foundation in Bergen (813558). Parts of this work have been done in the context of CEDAS, UiB's Center for Data Science. We are grateful as well to Juraj Pálenik, Bara Kozlíková, Bernhard Preim, Renate Grüner, and Eduard Gröller.



 Molecular
Molecular  Cellular
Cellular  Tissue
Tissue  Organ
Organ  Survey
Survey  Multiscale
Multiscale Analysis
Analysis Communication
Communication
 .
.  .
.  .
.  ,
,  ,
,  ,
,  ,
,  ,
,  .
.  , D471. doi:
, D471. doi:  .
.  ,
,  ,
,  ,
,  ,
,  ,
,  .
.  ,
,  ,
,  9.
9.
 ,
,  ,
,  ,
,  ,
,  ,
,  . doi:
. doi:  .
.  . Springer,
. Springer,  .
.  ,
,  ,
,  .
.  .
.  ,
,  ,
,  ,
,  , e0187341. doi:
, e0187341. doi:  ,
,  ,
,  .
.  .
.  ,
,  ,
,  .
.  ,
,  ,
,  ,
,  ,
,  ,
,  ,
,  .
.  .
.  ,
,  , 722. doi:
, 722. doi:  ,
,  .
.  , 3. doi:
, 3. doi:  .
.  .
.  ,
,  . doi:
. doi:  , W425–W430. doi:
, W425–W430. doi:  . The Eurographics Association,
. The Eurographics Association,  .
.  .
.  . doi:
. doi:  ,
,  ,
,  .
.  .
.  ,
,  .
.  ,
,  ,
,  .
.  .
.  ,
,  .
.  .
.  , e1005991. doi:
, e1005991. doi:  ,
,  ,
,  ,
,  ,
,  ,
,  ,
,  ,
,  ,
,  ,
,  ,
,  , 167351. doi:
, 167351. doi:  , eaag0025. doi:
, eaag0025. doi:  ,
,  ,
,  ,
,  ,
,  ,
,  .
.  . doi:
. doi:  ,
,  ,
,  ,
,  ,
,  ,
,  , e49774. doi:
, e49774. doi:  .
.  .
.  . doi:
. doi:  ,
,  ,
,  ,
,  .
.  ,
,  ,
,  .
.  .
.  .
.  ,
,  ,
,  ,
,  ,
,  ,
,  .
.  .
.  ,
,  ,
,  ,
,  .
.  ,
,  ,
,  .
.  ,
,  .
.  ,
,  .
.  .
.  ,
,  . Springer,
. Springer,  , 20190160. doi:
, 20190160. doi:  ,
,  . doi: 10.1109/TVCG.2020.3030336 14.
. doi: 10.1109/TVCG.2020.3030336 14.
 ,
,  ,
,  .
.  ,
,  , e61288. doi:
, e61288. doi:  . doi:
. doi:  ,
,  ,
,  ,
,  .
.  .
.  ,
,  ,
,  .
.  .
.  .
.  .
.  .
.  ,
,  .
.  .
.  ,
,  ,
,  ,
,  ,
,  ,
,  ,
,  .
.  .
.  ,
,  ,
,  . Springer Berlin Heidelberg,
. Springer Berlin Heidelberg,  .
.  .
.  .
.  ,
,  .
.  .
.  . Springer Berlin Heidelberg,
. Springer Berlin Heidelberg,  .
.  .
.  .
.  ,
,  .
.  ,
,  .
.  .
.  ,
,  .
.  ,
,  ,
,  . doi:
. doi:  .
.  ,
,  .
.  .
.  ,
,  ,
,  ,
,  ,
,  ,
,  .
.  ,
,  .
.  .
.  ,
,  .
.  ,
,  ,
,  .
.  ,
,  ,
,  ,
,  ,
,  .
.  ,
,  , e2001. doi:
, e2001. doi:  ,
,  . Springer,
. Springer,  ,
,  , 031710-1–031710-17. doi:
, 031710-1–031710-17. doi:  .
.  ,
,  ,
,  .
.  ,
,  .
.  ,
,  , 20120057. doi:
, 20120057. doi:  ,
,  .
.  , 12. doi: 10.389/fphys.2011.00012 20.
, 12. doi: 10.389/fphys.2011.00012 20.
 .
.  . doi:
. doi:  ,
,  ,
,  . Springer,
. Springer,  . doi:
. doi:  .
.  . Wilson Biodiversity Foundation,
. Wilson Biodiversity Foundation,  .
.  , 20170382. doi:
, 20170382. doi:  .
.  ,
,  . Gyldendal akademisk,
. Gyldendal akademisk,  ,
,  .
.  .
.  ,
,  . Springer-Verlag London,
. Springer-Verlag London,  . doi:
. doi:  ,
,  , 20150083. doi:
, 20150083. doi:  ,
,  ,
,  ,
,  , S56–S68. doi:
, S56–S68. doi:  ,
,  , dev177162. doi:
, dev177162. doi:  ,
,  .
.  .
.  .
.  ,
,  .
.  ,
,  ,
,  .
.  ,
,  ,
,  .
.  , S5–S15. doi:
, S5–S15. doi:  .
.  ,
,  .
.  ,
,  , S42–S55. doi:
, S42–S55. doi:  , S2–S4. doi:
, S2–S4. doi:  .
.  . Springer,
. Springer,  .
.  ,
, 
