Next-generation sequencing of endoscopically obtained tissues from patients with all stages of pancreatic cancer
Abstract
Routinely available clinical samples of all stages of pancreatic cancer are used in the present study to elucidate its molecular mechanisms and identify novel therapeutic targets. We evaluated the use of next-generation sequencing (NGS) of endoscopically obtained pancreatic cancer tissues. We enrolled 147 patients who underwent endoscopic ultrasound-guided fine-needle aspiration or endoscopic biopsy. The quantity and quality of the extracted DNA was assessed. Tissue samples were used for NGS of 78 cancer-related genes, from which gene alterations and microsatellite instability (MSI) were extracted. NGS was successful in 141 out of 147 (96%) cases. Gene alterations were detected in 134 out of 141 (91%) samples, among which eight out of 10 samples with a DNA concentration below the detection limit had some type of gene alteration. Targetable genes were detected in 28 (19.9%) cases. MSI and germline mutations in homologous recombination repair associated genes were detected in 5% and 3% of cases, respectively. Cox regression analysis revealed that metastasis (P < .005; hazard ratio [HR], 3.30) was associated with poor prognosis in all pancreatic cancer patients. In addition, fewer than three mutations (P = .03; HR, 2.48) and serum carcinoembryonic antigen levels >5 ng/mL (P < .005; HR, 3.94) were associated with worse prognosis in cases without and with metastasis, respectively. Targeted sequencing of all stages of pancreatic cancer using available samples from real clinical practice could be used to determine the relationship between gene alterations and prognosis to help determine treatment choices.
Abbreviations
-
- MMR
-
- mismatch repair
-
- NCC Oncopanel
-
- OncoGuide NCC
-
- NGS
-
- next-generation sequencing
1 INTRODUCTION
Pancreatic cancer is associated with poor prognosis and has a 5-year survival rate of <5% in the USA1 and 4.7% in Japan.2 There is an urgent need to elucidate the pathophysiology of the disease to identify early tumors and therapeutic targets to improve pancreatic cancer treatment. Next-generation sequencing (NGS) analysis of pancreatic cancer has revealed various genetic alterations (eg, chromosomal rearrangements, focal amplifications, and mutations) and deletions in many genes (eg, KRAS, TP53, CDKN2A, and SMAD4).3, 4 These comprehensive analyses have revealed the genetic landscape of pancreatic cancer and clarified its subtypes4 and genetic evolution; however, clinical application of these analyses is limited due to the frequent use of resected samples rather than clinically available biopsy samples. Furthermore, it is difficult to obtain a sufficient sequencing depth in tumor cell-poor tissues from clinically available samples.
Endoscopic ultrasound-guided fine-needle aspiration (EUS-FNA) and endoscopic biopsy of an invading tumor are the primary methods used to obtain tumor tissues from pancreatic cancer; however, these methods may not provide sufficient sample sizes. Recent advances in PCR methods using high fidelity DNA polymerases and NGS have enabled rapid, accurate, and comprehensive gene analysis to simultaneously detect multiple gene mutations and copy number variations (CNV) with high sensitivity, even using low amounts of DNA from clinical samples, such as formalin-fixed paraffin-embedded (FFPE) tissues.5-7 We previously used NGS to analyze 50 cancer-related genes from endoscopically obtained pancreatic cancer tissues8 and identified metastasis, serum carcinoembryonic antigen (CEA), and the number of gene alterations as prognostic markers. However, the study used a small cohort and excluded early-stage disease with insufficient gene regions to examine therapeutic targets.
In the present study, NGS was performed on endoscopically obtained FFPE samples from patients with early- and advanced-stage pancreatic cancer using our original gene panel, which covered a wide range of driver genes of pancreatic cancer and therapeutic targets. The gene panel was used to identify therapeutic targets and determine the clinical significance of these targets through comparisons with the patients’ clinical data.
2 MATERIAL AND METHODS
2.1 Tissue samples and ethics
We retrospectively evaluated 185 endoscopically obtained biopsy samples collected from 172 patients with pancreatic cancer who underwent EUS-FNA or endoscopic biopsy of an invading tumor at the Yamanashi University Hospital between July 2014 and December 2019. The EUS-FNA procedure was performed using 22- or 25-gauge needles of the normal, side hole, or Franseen type (EZ Shot 2 and EZ Shot 3 Plus, Olympus Medical Systems; Expect and Acquire, Boston Scientific). We punctured each sample 1-6 times (20 strokes/puncture) with negative pressure using a 20-mL syringe. Although rapid on-site cytologic evaluation cannot be conducted at our institution, we requested rapid cytologic evaluation at the Department of Pathology after one or two punctures to reduce the number of punctures. This retrospective study used residual samples of EUS-FNA specimens obtained for clinical diagnosis, and no additional puncture was performed for genetic analysis.
After eliminating duplicate samples from the patients and samples with failed NGS analysis or technical errors, 141 samples were finally analyzed by NGS (Figure 1A). A flow chart of the study is shown in Figure 1B. Tissue samples were obtained as 8-μm-thick sections derived from one or two FFPE blocks, and tumor components were separated from these sections using a Laser Capture Microdissection System (LCM, ArcturusXT, Life Technologies). A definite pathological diagnosis requires a certain amount of tumor cells or tumor area because small amounts of tumor-like cells appear similar to cells with degeneration caused by inflammation or mechanical damage. Thus, in samples with negative pathology, we excised the area where the tumor was presumed to be present because it could contain tumor cells.
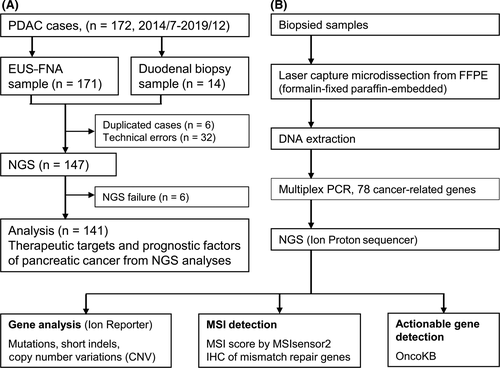
DNA was extracted from LCM specimens as previously described.5, 6 DNA from the biopsied specimens was extracted using GeneRead DNA FFPE Kits (QIAGEN) according to the manufacturer’s specifications. The quantity and quality of the extracted DNA was assessed using a NanoDrop instrument (Thermo Fisher) and Qubit platform (Thermo Fisher). The study was conducted in compliance with the principles of the Declaration of Helsinki and was approved by the Human Ethics Review Committee of Yamanashi University Hospital (Receipt number: 1326 and 1847). Written informed consent was obtained from all study participants. The study protocol was registered at UMIN Clinical Trials Registry (UMIN000044562).
2.2 Genetic mutational analysis of tissue samples using next-generation sequencing
Genetic analysis of the tumor specimens was performed by amplification of the extracted DNA (10 ng) using an Ion AmpliSeq Custom panel (Thermo Fisher) targeting 78 genes (Table S1) and containing 451 primer pairs. The custom panel differed from the ready-made panel used in our previous study.8 It was designed using the AmpliSeq Designer (Thermo Fisher) to cover pancreatic cancer driver genes and actionable targets listed in OncoKB.9 OncoKB is a knowledge base for precision medicine used to infer whether an existing molecular-targeted drug will be effective. Sequencing was performed with an Ion Chef System and an Ion Proton Sequencer (Life Technologies) using an Ion PI Hi-Q Chef Kit (Life Technologies). Data obtained in the present study were shared with the Japanese Genotype-Phenotype Archive (JGAS000315/JGAD000426).10
2.3 Identification of gene alterations and suspected microsatellite instability
Gene mutations and CNV were identified using Ion Reporter software version 5.10 (Thermo Fisher). Only mutations and CNV with a mutant allele frequency >4% (with a sequence read depth >100) and copy number >6 were considered truly present in the tissues to avoid false-positive variants due to sequencing errors. In the present study, the number of gene alterations was defined as the number of all mutations and CNV among the 78 cancer-related genes that were sequenced. Thus, all multiple mutations of the same gene and all additional mutations other than hotspot mutations in the COSMIC database were included. Detected gene alterations were then matched and classified as four levels according to OncoKB,9 which classifies genetic alterations based on actionability as follows: levels 1-3A, standard therapeutic intervention or compelling clinical evidence for the disease; level 3B, the presence of clinical evidence of another disease; and level 4, the presence of compelling biological evidence.
Microsatellite instability was detected with the MSIsensor2 tool,11 which uses machine learning methods to calculate the length distributions of microsatellites per site from sequence reads. The Ion AmpliSeq Custom panel used in the present study contained 20 homopolymer sites and three microsatellite sites consisting of a 2-6 base repetition in its target region. The microsatellite instability (MSI) score was calculated as the percentage of abnormal length distribution in the repeating sequences among total homopolymer or microsatellite sites. MSI was confirmed when the MSI score was ≥88, which was the cutoff value determined by a comparison of immunohistochemistry (IHC) of mismatch repair (MMR) genes (Figure S1).
2.4 Immunohistochemistry of mismatch repair genes
Anti–MLH1 (1:250 dilution; ab92312; AbCam plc), anti–MSH2 (1:8000 dilution; ab227941; AbCam plc), anti–MSH6 (1:500 dilution; ab92471; AbCam plc), and anti–PMS2 (1:100 dilution; ab110638; AbCam plc) antibodies were used as primary antibodies. IHC staining was performed according to the manufacturer’s instructions. Briefly, 3-μm-thick deparaffinized sections of FFPE were stained using primary antibodies specific for the above MMR genes. Antigens were retrieved by boiling tissue sections in Target Retrieval Solution (Dako). Envision+Dual Link HRP (Dako) was used as a secondary antibody and diaminobenzidine was used as the chromogen. IHC staining was blindly examined by two independent investigators.
2.5 Statistical analysis
Data that did not display a normal distribution were presented as the median (range) or median (interquartile range [IQR]). Factors associated with overall survival were identified using Cox multivariate regression analysis, in which P < .05 was considered significant. All statistical analyses of recorded data and graphic creations were performed using the Lifelines program (https://zenodo.org/badge/latestdoi/12420595) with the Python platform.
3 RESULTS
3.1 Patient characteristics and qualitative assessments of extracted DNA and next-generation sequencing
Among 147 samples that underwent NGS testing, 141 (96%) were successfully analyzed (Figure 1A); therefore, gene alterations in samples from 141 patients were analyzed with their clinical information. Table 1 shows the clinical characteristics of the 141 patients included in the study. Tumor stages were defined according to the TNM Classification of Malignant Tumors by the Union for International Cancer Control,12 and 4, 62, 16, and 59 patients in this study were classified as stages I, II, III, and IV, respectively. A total of 59 (42%) patients had stage IV pancreatic cancer and 82 (58%) received chemotherapy. Tissue samples were obtained by EUS-FNA or endoscopic biopsy in 129 (91%) and 12 (9%) cases, respectively (Table S2). A pathological diagnosis of malignancy was made using histology in 100 (71%) patients and cytology in 34 (24%) patients. However, a pathological diagnosis could not be made prior to therapy in seven (5%) patients, among whom three (2%) were diagnosed with malignancy using pathology of resected tissues, and four (3%) were diagnosed based on clinical course, respectively.
| Characteristics | PDC (n = 141) | |
|---|---|---|
| Age (years) | Median (range) | 70 (42-89) |
| Sex | Male/female | 81/60 |
| PDC location | Ph/Pbt | 85/56 |
| PDC size (mm) | Median (range) | 30 (11-75) |
| PDC stage | I/IIA/IIB/III/IV | 4/37/25/16/59 |
| Therapy | Operation/CRT/chemotherapy/BSC | 53/7/75/6 |
- Abbreviations: BSC, best supportive care; CRT, chemoradiation therapy; Pbt, pancreatic body and tail; PDC, pancreatic ductal carcinoma; Ph, pancreatic head.
The median (range) amounts of extracted DNA from the FFPE samples obtained by EUS-FNA or endoscopic biopsy were 20.6 (1-339) ng, except for 10 samples that had DNA concentrations below the limit of detection (LOD). In the NGS analyses of endoscopically obtained tissues, the target regions of the 78 cancer-related genes included 44 140 bases, with median (range) sequenced read depths of 4568 (448-29 865). Data for EUS-FNA and duodenal biopsies are shown in Table S2. The yield of extracted DNA (median [IQR]) was lower in EUS-FNA samples compared with that from duodenal biopsy (19.2 [10.2-34.2] ng vs 132.9 [79.7-202.7] ng, respectively; P < .001), whereas the number of samples with mutations in KRAS and any gene was not different between these two sets of samples (110/129 [85%], EUS-FNA vs 10/12 [83%], duodenal biopsy; P = 1.000) and (122/129 [95%], EUS-FNA vs 11/12 [92%], duodenal biopsy; P = .518) (Table S2).
3.2 Overview of genetic abnormalities of endoscopically obtained specimens in pancreatic cancer
Alterations to pancreatic cancer genes in the endoscopically obtained tissue samples were identified in 75 of the 78 analyzed cancer-related genes, and alterations of any type were detected in 134 cases (91%, Figure 2). Gene alterations were detected in eight of the 10 samples with extracted DNA concentrations below the LOD. The four most frequently altered genes in the tissue samples were KRAS (84%), TP53 (57%), SMAD4 (19%), and RET (18%), followed by SMO (16%), FGFR3 (14%), BRCA2 (10%), NF1 (10%), and CDKN2A (9%). When summarized according to signaling pathway, the Ras (KRAS, FGFR3, EGFR, ERBB2, FGFR1, ABL1, FGFR2, EGFR-AS1, FLT3, KIT, MET, PDGFRA, BRAF, RAF1, MEK, and ERK), mTOR (KRAS, RET, FGFR3, PIK3CA, MTOR, PTEN, ERBB2, FGFR1, ABL1, FGFR2, ALK, KIT, MET, PDGFRA, NRAS, AKT1, MTOR-AS1, ERGR, and PI3K), cell cycle (TP53, CDKN2A, and RB1), JAK/STAT (ERBB2, ABL1, ALK, FLT3, JAK2, JAK2, and ERGR), and Wnt (APC, CTNNB1, and CDH1) signaling pathways were activated by their constituent gene alterations in 89%, 88%, 62%, 11%, and 9% of samples, respectively.
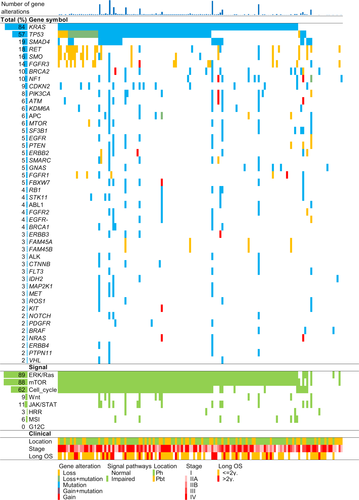
3.3 Actionable genes in pancreatic cancer
We next matched the detected gene alterations with OncoKB, which classifies genetic alterations into four levels according to an actionability scale. Oncogenic mutations in KRAS were most frequently detected in the cohort, and they were associated with resistance to molecular targeted drugs, such as cetuximab and panitumumab (Table S3). The next most frequently detected actionable genes were BRCA2 and PIK3CA, which were associated with the efficacy of talazoparib, olaparib, rucaparib, and fulvestrant plus alpelisib. In total, 19.9% of patients with gene alterations corresponded to levels 1-2 of OncoKB (Table S4). Furthermore, germline mutations in homologous recombination repair (HRR)-associated genes, which are efficacy markers for olaparib,13 were detected in 3% of patients. None of the patients had a G12C mutation in KRAS, a specific target for sotorasib,14 and MSI, which is an efficacy marker for immune checkpoint inhibitors,15 was detected in 5% of the patients (Figure 2).
3.4 Genetic diagnosis of malignancy in pathologically negative cases
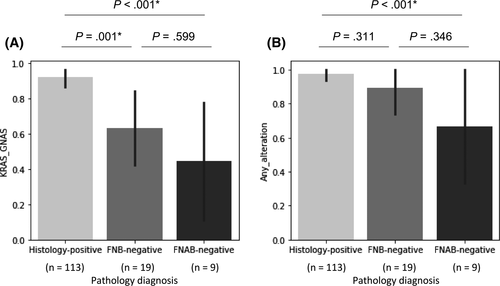
Pathological diagnoses of endoscopically obtained samples were classified into three groups: histology-positive, with histological malignancy findings using EUS-FNB or duodenal biopsy (n = 113, 80.1%); FNB-negative, with negative findings using EUS-FNB but positive findings using EUS-FNA cytology (n = 19, 13.5%); and fine-needle aspiration biopsy (FNAB)-negative, with negative findings using EUS-FNB and EUS-FNA cytology (n = 9, 6.4%). Two, three, and four of the nine FNAB-negative patients were diagnosed with malignancy using cytological testing of bile juice or ascites, pathology of surgical specimens, and imaging and bad clinical course, respectively. Patients who were pathologically negative using either EUS-FNB or EUS-FNA cytology had gene alterations in KRAS or GNAS and at least one of 78 genes in four (44%) and six (67%) of the nine cases, respectively, suggesting the diagnostic ability of genetic analysis even in FNAB-negative cases (Figure 3).
3.5 Genetic and clinical factors associated with overall survival
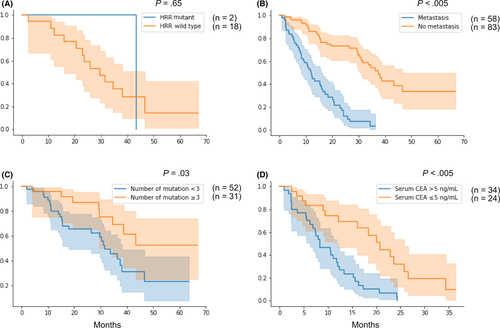
Survival analysis was performed because the relationship between the genetic and clinical factors, including overall survival, remains poorly understood. We first analyzed the survival rates of patients treated using FOLFIRINOX (n = 20) according to HRR gene status because HRR status is related to the therapeutic effect of the platinum-containing regimen. We revealed a tendency toward better survival in cases with HRR mutations (Figure 4A). We next performed Cox regression analysis for overall survival with genetic and clinical factors (Table 2) and revealed that the presence of metastasis (P < .005; hazard ratio [HR], 3.30; Figure 4B) and serum CEA levels >5 ng/mL (P = .01; HR, 1.89) were associated with worse prognosis. Because metastasis was found to have a strong influence on overall survival, we then performed Cox regression analysis by stratifying metastasis and revealed that the number of mutations <3 (P = .03; HR, 2.48, Figure 4C) and serum CEA levels >5 ng/mL (P < .005; HR, 3.94; Figure 4D) were associated with worse prognosis in cases without and with metastasis, respectively (Table 3). We also stratified patients into those who underwent surgical resection from the viewpoint of clinical common sense and then performed Cox regression analysis (Table S5). No factors influenced the prognosis in surgically treated cases, whereas metastasis (P < .005; HR, 4.16) and CEA levels >5 ng/mL (P < .005; HR, 2.28) were associated with poor prognosis in patients who did not undergo surgical resection.
| Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|
| n (total =141) | P-value | HR (95%CI) | P-value | |
| Age ≥70 years | 72 | .22 | ||
| Female sex | 60 | .12 | ||
| Non–operative therapy | 88 | <.01** | 1.76 (0.86-3.63) | .12 |
| Located in pancreas head | 85 | .23 | ||
| Size >20 mm | 113 | .01* | 1.76 (0.89-3.47) | .11 |
| Stage IV (With metastasis) | 58 | <.01** | 3.30 (1.68-6.48) | <.005** |
| CA19-9 > 100 U/mL | 71 | <.01** | ||
| CEA >5 ng/mL | 56 | <.01** | 1.89 (1.11-3.04) | .01* |
| MSI | 7 | .14 | ||
| Number of indels ≥2 | 8 | .09 | ||
| Number of mutations <3 | 46 | .02* | 0.69 (0.40-1.19) | .19 |
| Alteration in genes | ||||
| KRAS | 119 | .93 | ||
| TP53 | 80 | .79 | ||
| SMAD4 | 27 | .20 | ||
| RET | 25 | .07 | ||
| SMO | 22 | .02* | 1.32 (0.73-2.37) | .35 |
| FGFR3 | 20 | .49 | ||
| NF1 | 14 | .29 | ||
| BRCA2 | 14 | <.01** | 0.43 (0.12-1.56) | .2 |
| Altered signaling pathways | ||||
| ERK/Ras | 125 | .84 | ||
| mTOR | 124 | .56 | ||
| Cell cycle | 87 | .73 | ||
| Wnt | 12 | .48 | ||
| JAK/STAT | 15 | .33 | ||
| HRRa | 18 | .04* | ||
- CI, confidence interval; HR, hazard ratio; HRR, homologous recombination repair; MSI, microsatellite instability.
- a Somatic and germline mutation.
- * P < .05.
- ** P < .01.
| Stage I-IIIa (n = 83) | Stage IVb (n = 58) | |||
|---|---|---|---|---|
| HR (95%CI) | P-value | HR (95%CI) | P-value | |
| Non–operative therapy | 1.72 (0.81-3.67) | .16 | 2.65 (0.35-20.3) | .35 |
| Size >20 mm | 1.63 (0.66-4.11) | .28 | 2.99 (1.00-8.96) | .05 |
| CEA >5 ng/mL | 0.95 (0.42-2.16) | .91 | 3.94 (1.93-8.04) | <.005** |
| Number of mutations <3 | 2.48 (1.09-5.63) | .03* | 1.47 (0.71-3.04) | .3 |
- CI, confidence interval; HR, hazard ratio.
- a No metastasis.
- b With metastasis
- * P < .05.
- ** P < .01.
4 DISCUSSION
The present study evaluated the use of NGS analysis of endoscopically obtained pancreatic cancer tissue samples using the original gene panel that targeted 78 cancer-related genes with driver genes of pancreatic cancer and therapeutic targets. NGS analysis was successful in 141 of 147 (96%) clinically obtained samples, even with small amounts of FFPE samples, and therapeutic targets were detected in 19.9% of cases. Furthermore, clinical and genetic factors that were associated with prognosis were identified by analyzing patients with all stages of pancreatic cancer.
Endoscopically obtained pancreatic cancer samples could be used for genetic analysis to identify therapeutic targets and factors associated with prognosis. Recent commercially available gene panel tests, such as FoundationOne CDx (F1CDx)16 and OncoGuide NCC (NCC Oncopanel),17 have been widely used and are covered by national health insurance for all patients with recurrent and refractory solid tumors in Japan,18 promoting the spread of personalized medicine. However, FoundationOne CDx and OncoGuide NCC Oncopanel Systems require ≥50 and ≥200 ng of DNA, respectively, which is not easy to obtain by endoscopic biopsy, especially EUS-FNA. Previous studies reported successful NGS using samples obtained by EUS-FNA targeting lung cancer19 as part of the MSK-IMPACT study,20 with a success rate of 86% (99 of 115 cases). The reasons for NGS failure were low DNA yield (<50 ng), technical failure, and insufficient tumor content. In the present study, NGS analysis was successful in 141 out of 147 (96%) endoscopically obtained samples, even with small amounts (<10 ng) of DNA. Although we did not analyze the factors associated with NGS success in detail, positivity in rapid pathological diagnosis during EUS-FNA appeared to be associated with its success (data not shown).
Genetic analyses should be performed even using small amounts of clinically obtained DNA to apply precision medicine for pancreatic cancer. An abundance of interstitial pancreatic cancer tissue can hinder the detection of gene alterations by extensive gene analysis with insufficient sequence depths, particularly when the tissue samples are very small in size. Therefore, targeted sequencing of endoscopically obtained FFPE samples using a compact panel with 78 cancer-related genes, as performed in the present study, is significant for clinical practice. Our original panel using endoscopically obtained FFPE samples could be used to detect targetable gene alterations corresponding to OncoKB level 1 or 2 (defined as targetable markers for drugs that were currently available in clinical practice)9 in 19.9% of cases. Furthermore, none of the cases had a G12C mutation in KRAS, which could be a specific target for Sotorasib.14 In contrast, MSI, which could be an efficacy marker for immune checkpoint inhibitors,15 was detected in 5% of cases, and a germline mutation and germ and/or somatic mutation in the HRR gene, which is related to the therapeutic effect of a platinum-containing regimen,21 were detected in 3% and 15% of cases, respectively. Although there were some differences, the detection rates of these targetable genes were similar to those previously reported.
Our findings are novel in that they revealed a relationship between overall survival and clinical factors, including genetic alterations in endoscopically obtained samples from a cohort of pancreatic tumors of various stages. Notably, our cohort included resectable as well as unresectable pancreatic cancers, whereas most published data on genetic analysis of pancreatic cancer is from resected tumor samples.8, 22 Thus, our findings are valuable and contribute to understanding the relationship between genetic alterations and features of all stages of pancreatic tumors because our analysis revealed that a poor prognosis was related to metastasis and high CEA levels as well as a low number of mutations in patients without metastasis.
The reasons for the association of the presence of ≥3 mutations and better prognosis in patients with stage I-III lesions remain unknown. To better elucidate the relationship between the number of mutations and prognosis, we conducted post hoc analysis in subgroups classified by age, sex, therapy, tumor location, tumor size, and tumor stage (Figure S2), among which the presence of ≥3 mutations was associated with better prognosis only in stages I-III. Although there were no similar reports in pancreatic cancer, many studies have stated that patients with a high number of mutations or MSI-high status, which leads to hypermutation, had better survival in gastric cancer after curative resection,23 stage III colon cancer,24 curatively resected stage II colon cancer,25 and stage IIIA non–small-cell lung cancer.26 Although the reason why the number of mutations was related to patient survival remains unclear, we presume that a high number of mutations derived from microsatellite instability is associated with lymphocytic reaction, including tumor-infiltrating lymphocytes in colorectal carcinoma27 and pancreatic cancer28 low-grade pathology, which could permit curative therapy in stages I-III. In addition, although platinum-based chemotherapy was reported as effective in patients with somatic hypermutation in ovarian cancer,29 no association between the number of mutations and patient survival has been identified in pancreatic cancer.30, 31 We wanted to analyze the relationship between the number of mutations and the efficacy of platinum-based therapy in patients with stage IV lesions, but this was precluded by the insufficient number of cases.
Our study findings have several clinical implications. First, we managed to overcome the challenges of obtaining pancreatic cancer samples. Outsourced genomic analyses require approximately 50-200 ng of DNA, which is not easily obtainable in real practice. In the present study, a median of 19.2 and 132.9 ng of DNA was obtained from EUS-FNA and duodenal biopsies, respectively, and was used to achieve the same detection rate of gene alterations (Table S2). Moreover, gene alterations were detected in eight out of 10 cases using DNA concentrations below the LOD. Second, our gene analysis data can be used to derive important information regarding prognosis and treatment selection.
The present study has several limitations. First, the design was retrospective and used data obtained from a single center. Second, we were unable to calculate the tumor content of the obtained tissue samples; therefore, samples in which NGS analysis failed may have had a low tumor content.
In conclusion, we performed targeted sequencing of endoscopically obtained pancreatic cancer FFPE samples that were available in real clinical practice and evaluated the relationship between gene alterations and prognosis to help determine treatment choices. Our findings may be used to improve the clinical outcomes of patients with pancreatic cancer.
ACKNOWLEDGMENTS
This study was supported by grants from the Japan Society for the Promotion of Science (JSPS KAKENHI grant numbers: 18K07999 and 19K08418; http://www.jsps.go.jp/j-grantsinaid/). We thank Takako Ohmori and Tomoko Nakajima for their valuable technical assistance, Hiroko Amemiya for her secretarial assistance, and Masato Takano (Orange Square) for his computer programming assistance. The authors would like to thank Enago (www.enago.jp) for the English language review.
DISCLOSURE
The authors have no potential conflicts of interest to disclose.




