Not all epidermal growth factor receptor mutations in lung cancer are created equal: Perspectives for individualized treatment strategy
Funding Information
Grant-in-Aid for Scientific Research (Grant/Award Number: ‘16K19989’).
Abstract
Somatic mutations in the epidermal growth factor receptor (EGFR) gene are present in approximately 20% (in Caucasians) to 40% (in East Asians) of adenocarcinomas of the lung. Targeted therapy for these lung cancers has been established based on evidence regarding mainly common mutations; that is, exon 19 deletions (Del19) and L858R. EGFR-tyrosine kinase inhibitors (TKI), gefitinib, erlotinib or afatinib showed high objective response rates (ORR) of approximately 60%. Several studies suggested that Del19 might be more sensitive to EGFR-TKI than L858R. On the other hand, it has been difficult to establish evidence for other less common mutations, accounting for 12% of all EGFR mutations, because there are many variants and many studies have excluded patients with these uncommon mutations. However, recent studies revealed that these rare genotypes could be targetable if appropriate TKI are selected. For example, G719X (X denotes A, S, C and so on), Del18, E709K, insertions in exon 19 (Ins19), S768I or L861Q showed moderate sensitivities to gefitinib or erlotinb with ORR of 30%–50%. However, afatinib appeared to be especially effective for these tumors. Although Ins20s (except for insFQEA) have been regarded as resistant mutations, osimertinib may be effective for rare subtypes of them and nazartinib (EGF816) is promising for the majority of them. For the further development of targeted therapy in all EGFR mutations, it is important to precisely detect targetable mutations, to select the most appropriate TKI for each mutation, and to continue investigating in vitro studies and collecting clinical data on even rare mutations.
Somatic mutations in the kinase domain of the epidermal growth factor receptor (EGFR) gene are detected in approximately 40% and 17% of lung adenocarcinoma in Asians1 and in Caucasians,2 respectively. When these biomarkers were first developed, early studies simplified the complexity of tumor genotype by dichotomizing them as mutant or wild type. Fortunately, common mutations (i.e. exon 19 deletions [Del19] and L858R mutation in exon 21) are associated with sensitivity to EGFR tyrosine kinase inhibitors (TKI).3, 4 Targeted therapies for these lung cancers were established based on 7 phase III randomized trials.5-11
EGFR mutations other than Del19 and L858R are variably termed either minor (less common) or uncommon mutations. The need for appropriate management of patients with these uncommon mutations is increasing because the incidence of uncommon EGFR mutations is comparable to rare targetable driver genes such as ROS1 and RET.12-15 We recently reported that second generation EGFR-TKI, afatinib or neratinib, are especially effective for EGFR exon 18 mutations compared with other EGFR-TKI, indicating the significance of mutation-specific EGFR-TKI selection.16
In this review, we comprehensively collected data on the frequency, in vitro sensitivity and treatment response of lung cancers harboring common and uncommon EGFR mutations to provide insight for the future direction of rational therapeutic strategy .
EGFR Pathway and Mutations in the EGFR
EGFR is one of the ERBB family receptor tyrosine kinases that consists of four members: EGFR (also known as ERBB1/HER1), ERBB2/HER2/NEU, ERBB3/HER3 and ERBB4/HER4. Specific ligands bind to the extracellular domain of EGFR, which leads to the formation of homodimers and heterodimers. Dimerization stimulates intrinsic tyrosine kinase activity of the receptors and triggers the autophosphorylation of specific tyrosine residues. Signal transducers initiate multiple downstream pathways such as MAPK, PI3K-AKT and STAT 3 and 5, which regulate proliferation and apoptosis.17
The EGFR gene, located on chromosome 7p12, consists of 28 exons and 27 introns. In 2004, somatic mutations in the kinase domain were discovered in patients with lung cancer whose tumor responded to gefitinib.3, 4 EGFR mutations shift the equilibrium of protein structures from an inactive state into an active state, resulting in the increased and sustained phosphorylation of EGFR and other HER family proteins without ligand stimulation.18
Types of EGFR Mutations According to the COSMIC Database
The catalogue of somatic mutations in cancer (COSMIC) is the largest open access database.19 As of May 2016, approximately 16 000 EGFR mutations are registered. According to this database, as many as 594 types of EGFR mutations are reported. Among them, 93% are present in the first four exons (18–21) of the gene encoding tyrosine kinase domain. Although COSMIC is extremely useful for comprehensive overview of EGFR mutations, including rare mutations, the results should be interpreted cautiously because the database consists of various data. For example, there was a discrepancy in the frequency of Del19 and L858R in conventional published data.20 Del19 accounts for approximately half of L858R (Table 1).
| Category | Present survey | COSMIC (n = 16138) |
|---|---|---|
| Del19 | 44.8 | 27.4 |
| L858R | 39.8 | 52.7 |
| Ins20 | 5.8 | 2.0 |
| G719X | 3.1 | 2.8 |
| S768I | 1.1 | 0.9 |
| L861Q | 0.9 | 1.8 |
| Ins19 | 0.6 | 0.2 |
| E709X | 0.3 | 0.5 |
| Del18 | 0.3 | 0.1 |
| Others | 3.3 | 5.0 |
| T790M | Excluded | 6.6 |
| Total (%) | 100 | 100 |
- EGFR, epidermal growth factor receptor.
Frequency of EGFR Mutations by Compilation of Recent Large Studies
Three factors appear to complicate estimations of the true frequencies of each EGFR mutation in clinics: methods for detecting mutations, the presence of complex mutations and publication biases.
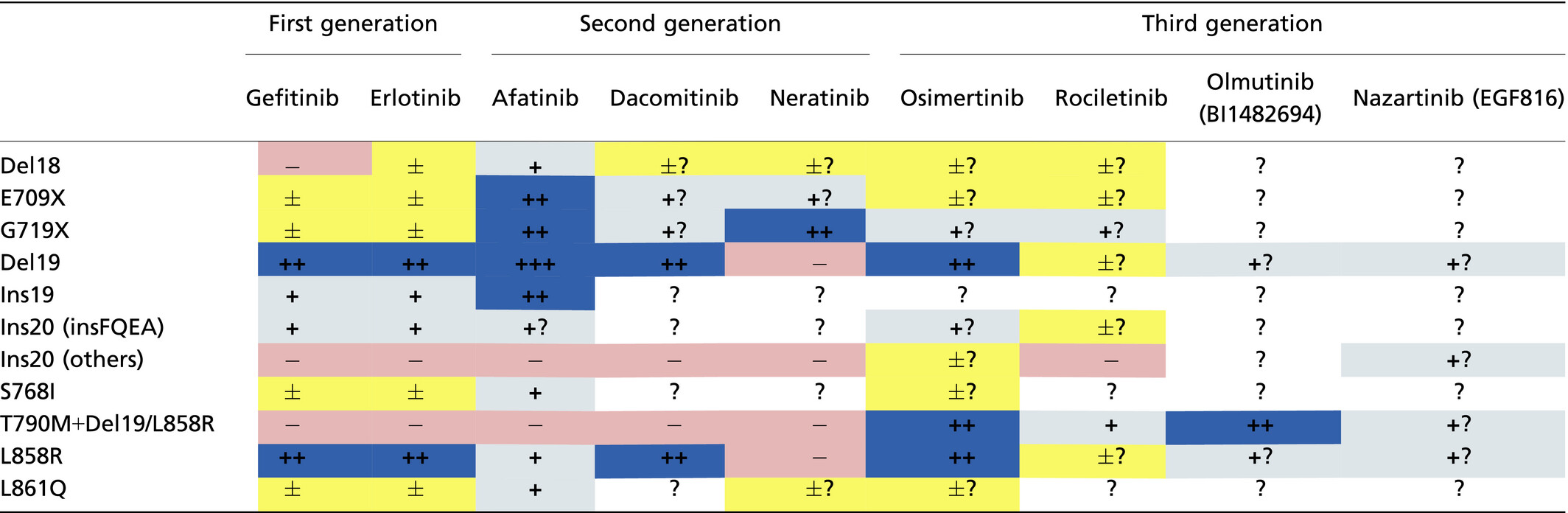
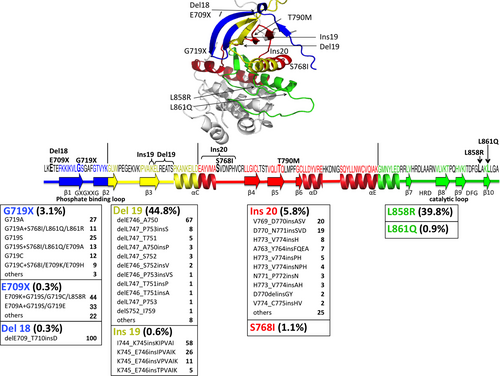
Sanger sequencing has been performed to detect mutations throughout the exons sequenced (usually exons 18–21), although the sensitivity is relatively low (requiring approximately 10% of the mutation allele).21 Next-generation sequencing can also achieve broad mutation detection. Importantly, rare mutations may include artifactual mutations that are generated during the pre-analytic period.22 In contrast, mutation-specific diagnostic kits have been developed for rapid and easy testing in clinical settings. Therascreen (Qiagen, Manchester, UK) and cobas (Roche, Basel, Switzerland) are approved by health authorities as in vitro diagnostic kits. These assays can detect the following specific mutations with high sensitivities (requiring approximately 1% of the mutation allele): G719A/S/C, Del19, S768I, exon 20 insertions (Ins20: V769_D770insASV, D770_N771insG/SVD and H773_V774insH), T790M, L858R and L861Q (Fig. 1). In other words, there is no way for other mutations to be detected. Although using these diagnostic kits is the standard method for detecting EGFR mutations in clinical practice, it is necessary to improve them to be able to detect rare but targetable mutations.
Multiple EGFR mutations are sometimes detected in the same tumor and these mutations have been referred to as co-mutations, complex mutations or compound mutations.23-26 Numeration for these mutations is not defined: some studies include them as a part of the representative mutation, such as Del19 or L858R, and others count these mutations independently (i.e. double-counting).
Oxnard and Jänne provide insightful comments on publication biases. Not all data on specific genotypes reaches the published literature: common genotypes are often included in prospective trials; less common or rare genotypes may be described in observational series or case reports; and the completeness of the data in meta-analyses inherently depends upon selection criteria as well as publication biases.27
Ideally, prospective large studies using the same method can clarify the actual frequency. Considering these factors as possible, we collected large studies conducted by single institutions or multi-institutional studies using the same protocol (Fig. 1). Subsequently, we focused on rare but targetable subsets including Ins19,28 Del1829 and E709X.29 Pretreatment T790M was excluded from our survey because the majority of them exist as complex mutations and the frequency vary widely from 3.9% to 64% based on the sensitivities of assays.30 In addition, the frequency of germline T790M remain unclear.31-33
First, Second and Third Generation EGFR -Tyrosine Kinase Inhibitors
Gefitinib and erlotinib, first generation (1G) EGFR-TKI, reversibly bind to the ATP-binding pocket of EGFR. Randomized phase III trials have demonstrated the superiority of these TKI, in terms of progression free survival (PFS), to conventional chemotherapy in patients with lung cancers harboring EGFR mutations.5, 6, 11 However, these TKI inevitably acquire resistance after an initial response. The secondary mutation T790M accounts for approximately 50%–60% of acquired resistance to 1G-TKI.34, 35
Irreversible pan-HER (EGFR, HER2 and HER4) TKI, so-called second generation (2G) TKI, were developed to overcome the T790M mutation. Despite the promising preclinical data, clinically available concentrations of the drug did not reach the treatment range for T790M tumors because of relatively severe adverse events compared with 1G-TKI due to the inhibition of wild type EGFR. However, afatinib has been approved as the first-line treatment for patients with EGFR-mutant lung cancers based on phase III trials.9, 10 Dacomitinib had a high objective response rate (ORR) of 76% in a phase II trial and continues to undergo clinical evaluation.36 Neratinib is also one of the 2G-TKI. However, its development for lung cancer was abandoned because it was not effective for common EGFR-mutant tumors, although it was effective for G719X tumors.37
The pyrimidine-based third generation (3G) TKI have been developed targeting T790M as well as common mutations without inhibiting wild-type EGFR.38, 39 Osimertinib has been approved for T790M tumors based on the high ORR of approximately 60% for tumors with T790M mutations as a resistance mechanism of 1G-TKI.38 C797S secondary mutation was detected in T790M-positive tumors that acquired resistance to osimertinib.40 Furthermore, C797S mutation appeared to be sensitive to 1G-TKI, and even C797S+T790M in trans can be treated with a combination of 1G and 3G-TKI.41 In contrast, the development of rociletinib was abandoned because the initially reported ORR of 59% was reduced to 45%: initial data were not unconfirmed partial responses although partial responses must be maintained on a second scan obtained at least 4 weeks later.42 Recently, olmutinib (BI1482694/HM61713) was approved for T790M-positive tumors in South Korea and received FDA breakthrough therapy designation.43 Other 3G-TKI, nazartinib (EGF816) and ASP8273, are undergoing clinical evaluation.44
Treatment Strategy by Mutation-specific Tyrosine Kinase Inhibitors Selection
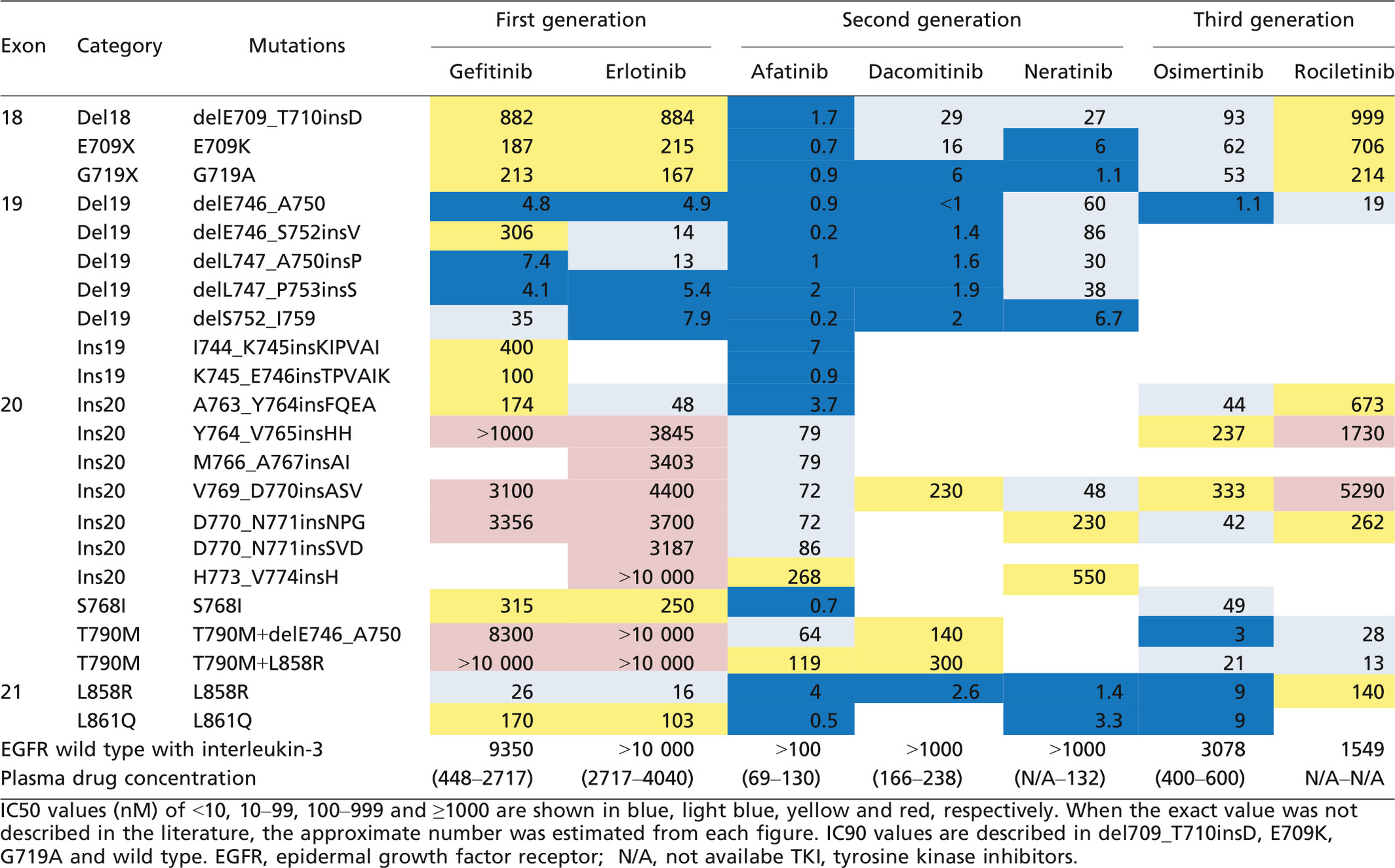
When we discuss treatment strategies for heterogeneous EGFR populations, biases should be considered, especially for the data on less common or rare mutations. To compensate for the weak evidence for such mutations, we also collected data on in vitro sensitivities using Ba/F3 cells (Table 2) as well as clinical response to TKI (Table 3). Notably, the murine pro-B cell line Ba/F3 depends on interleukin-3 (IL-3) for its survival and growth. Accordingly, the growth of Ba/F3 cells transfected with specific EGFR mutation in the absence of IL-3 indicates oncogenic ability, which can exclude artifactual mutations. Of course, the methodological differences and clinically available concentrations should be considered in the interpretation of in vitro sensitivities.
| Category | Mutation | CR | PR | SD | PD | ORR |
|---|---|---|---|---|---|---|
| Del18 (n = 4) | delE709_T710insD | 1 | 1 | 2 | 25 | |
| E709X (n = 15) | E709A/H/K+complex | 8 | 5 | 2 | 53 | |
| E709K+G719A/C/S/L858R | 5 | 1 | 1 | 71 | ||
| E709A+G719A/C/S/L858R | 3 | 4 | 43 | |||
| E709H+T710del | 1 | 0 | ||||
| G719X (n = 202) | G719X/A/S/C | 48 | 49 | 51 | 32 | |
| G719X | 36 | 38 | 33 | 34 | ||
| G719A | 8 | 6 | 10 | 33 | ||
| G719S | 3 | 3 | 7 | 23 | ||
| G719C | 1 | 2 | 1 | 25 | ||
| G719X/A/S/C+complex | 1 | 33 | 18 | 6 | 59 | |
| G719X+S768I/L861Q | 13 | 6 | 68 | |||
| G719A+E709A/K/S720F/T725M/L747S/S768I/L833V+V834C/L858R/L861Q/R | 1 | 10 | 5 | 3 | 58 | |
| G719S+Q701L+I706T/E709A/K/S768I/L858R/L861Q | 9 | 6 | 3 | 50 | ||
| G719C+E709K/K714N | 1 | 1 | 50 | |||
| Ins19 (n = 10) | Ins19 | 4 | 6 | 40 | ||
| I744_K745insKIPVAI | 4 | 4 | 50 | |||
| K745_E746insIPVAIK | 2 | 0 | ||||
| Ins20 (n = 59) | Ins20 | 10 | 14 | 35 | 17 | |
| D770_N771insSVD | 1 | 3 | 9 | 8 | ||
| V769_D770insASV | 1 | 3 | 6 | 10 | ||
| A763_Y764insFQEA | 6 | 1 | 86 | |||
| H773_V774insH | 2 | 3 | 0 | |||
| Y764_V765insHH | 1 | 0 | ||||
| M766_A767insASV/insWPA | 1 | 1 | 0 | |||
| A767_S768insSVR | 1 | 0 | ||||
| V769_D770delinsGI | 1 | 0 | ||||
| D770_N771insGL/insGT/delinsGY | 1 | 5 | 0 | |||
| N771_P772insN/delinsG/delinsSY/delinsKPP | 1 | 1 | 2 | 25 | ||
| P772_H773insDNP/insYNP/dupPH/P772_V774dupPHV/P772_C775dupPHVC | 1 | 4 | 20 | |||
| H773_V774insAH/insNPH | 1 | 3 | 0 | |||
| S768I (n = 30) | S768I | 5 | 2 | 5 | 42 | |
| S768I+G719X/G724S/V769L/V774M | 9 | 8 | 1 | 53 | ||
| L861Q (n = 76) | L861Q | 25 | 24 | 15 | 39 | |
| L861Q+G719X | 11 | 1 | 92 |
- CR, complete response; EGFR, epidermal growth factor receptor; ORR, objective response rate; PD, progressive disease; PR, partial response; SD, stable disease; TKI, tyrosine kinase inhibitors.
Common mutations Del19 and L858R
Del19 and L858R account for 44.8% (2573/5741) and 39.8% (2283/5741) of EGFR mutations, respectively.29, 45-48 Evidence of these common mutations has been developed in prospective trials: gefitinib, erlotinib and afatinib showed ORR of approximately 60% and PFS of 9–13 months.5-11 To clarify the appropriate TKI selection, efficacies of several EGFR-TKI have been directly compared in prospective trials. In previously treated patients, PFS was not significantly different between patients treated with gefitinib and those with erlotinib in WJOG5108L study:49 The LUX-lung 7 trial showed the superiority of afatinib compared to gefitinib as the first-line treatment in terms of PFS with a hazard ratio (HR) of 0.73 (95% CI 0.57–0.95).50 Currently, ARCHER1050 (dacomitinib versus gefitinib), FLAURA (osimertinib versus gefitinib or erlotinib) and TIGER1 (rociletinib versus erlotinib) trials are ongoing.
Conventionally, Del19 and L858R have been classified into one sensitive group. However, a meta-analysis including seven randomized trials, which compared EGFR-TKI to platinum doublet chemotherapy, was conducted to compare the HR of PFS between the Del19 group and the L858R group. The study revealed that the HR of PFS for tumors with Del19 was 50% greater (HR 0.24, 95% CI 0.20–0.29) than for tumors with L858R (HR 0.48, 95% CI 0.39–0.58).51 LUX-lung 3 and 6 studies showed a survival benefit of afatinib in patients with Del19-tumors but not for those with L858R-tumors.52 These data suggested that even these common mutations have different chemosensitivities. We recommend afatinib for first-line treatment in patients with Del19 tumors. Mature data on the overall survival in LUX-lung 7 trial should be considered to discuss the first line treatment for L858R tumors. Interestingly, Chen et al. reported that pretreatment T790M was more frequent in L858R-tumors than in Del19-tumors, although the differences were observed only in studies using methods with a detection limit <5%.30 However, when background mutations in tumors with acquired resistance by T790M were examined, Del 19 was more common than L858R. The number of patients with Del19 + T790M tumors who enrolled in phase I/II trials for osimertinib38 and rociletinib39 was approximately twice as large as the number of patients with L858R+T790M.
Del19 includes at least 30 variants.53 Deletion starting at E746 (the majority of them are delE746_A750), E747 and others are present in 73% (272/373), 25% (92/373) and 2% (9/373), respectively.53-55 Rare variant delE746_S752insV may be less sensitive to gefitinib according to the in vitro data (Table 2).56-58 The clinical data are controversial: the largest study by Chung et al. reported that ORR to 1G-TKI in Del starting at E746 were lower than those in Del starting at L747 (68.2% and 79.6%, respectively),53 whereas other groups showed the opposite tendency.54, 55 Notably, ORR in 7 patients with Del starting at 748, 751 or 752 was only 43%.53
Ins 20
Insertional mutations in exon 20 (Ins 20) account for 5.8% (134/2307) of all EGFR mutations and, based on our survey, consist of 44 types of mutations (Fig. 1).59-62 Inserted residues seem to be a part of the wild-type sequence; thus, these mutations may be referred to as duplications. As mentioned above, only four types are detectable using approved diagnostic kits, accounting for only 49% (66/134). In addition, Yasuda et al. reported an additional 31 types of mutations.18, 63
In a compilation of data on the treatment response to 1G-TKI, ORR was only 17% (10/59) (Table 3).61-65 Even 2G-TKI achieve ORR of only 10%.18, 66 However, osimertinib may be effective for the rare subtype D770_N771insNPG.67 Jia et al. (2016) reveal that one of the 3G-TKI, nazartinib (EGF816), has promising activity in overcoming the major subtypes V769_D770insASV and D770_N771insSVD.68 Of note, A763_Y764insFQEA was sensitive to 1G-TKI with ORR of 86%. Thus, this mutation, accounting for 7% of Ins20, should not be overlooked in clinical practice.63, 67
G719X, Del18 (delE709_T710insD) and E709X
These genotypes are present in 3.1% (100/3186), 0.3% (9/3186) and 0.3% (9/3186), respectively (Fig. 1).29, 61, 69 Although G719X includes many variants, 97% of them are G719A/S/C which can be detected by diagnostic kits. Approximately one-third of G719X mutations are present as complex mutations and they tend to be in combination with S768I or L861Q (Fig. 1). Most E709X present as complex mutations and the paired mutations tend to be G719X or L858R. Accordingly, patients with E709X tumors are, fortunately, found to have at least the representative G719X or L858R using diagnostic kits. However, Del18 can be missed.
G719A, E709K and Del18 appeared to have high sensitivity to afatinib or neratinib compared to 1G or 3G-TKI in our in vitro study.29 ORR to 1G-TKI in patients with G719X-tumors as a single mutation and Del18 were only 32% and 25%, respectively (Table 3).29, 61, 69-73 Relatively high ORR of 53% (for E709X) and 59% (for G719X) were observed in patients with complex mutations. Combined analysis of LUX-lung 2, 3 and 6 trials demonstrated that ORR for G719X tumors treated with afatinib was 78% (14/18).66 In addition, two out of four patients with tumors harboring E709X+G719X or L858R, as well as our patient with Del18-tumor, achieved partial responses to afatinib after the treatment with 1G-TKI.29, 74
S768I and L861Q
S768I and L861Q account for 1.1% (39/3712) and 0.9% (34/3712), respectively (Fig. 1).46-48 These mutations can be coupled with G719X but the actual frequency of the complex mutation remains uncertain.
ORR to 1G-TKI in S768I and L861Q tumors were only 42 and 39%, respectively (Table 3).26, 29, 61, 69-73, 75-80 Our in vitro data showed that both mutations are sensitive to afatinib. In addition, osimertinib may be effective for L861Q tumors.81 Combined analysis of LUX-lung 2, 3 and 6 trials reported that ORR for S768I or L861Q tumors treated with afatinib was 100% (8/8) and 56% (9/16), respectively. However, only 1 patient had S768I as a single mutation, and the remaining 7 patients had S768I+G719X or L858R. On the other hand, half of the patients had L861Q as a single mutation. Therefore, further clinical data should be collected to confirm these sensitivities.
Ins 19
Ins19 is a subset accounting for 0.6% (26/4519).28, 48, 82, 83 I744_K745insKIPVAI is the common type of insertion. There are a few variants of insertion starting at K745. He et al. suggested that these genotypes are sensitive to 1G-TKI and afatinib.28 Although only limited data are available, ORR to 1G-TKI is 40%.28, 82-84 One patient with K745_E746insIPVAIK tumor achieved partial response to afatinib.28
Conclusions
EGFR mutations in lung cancer are extremely complicated and each mutation appears to have unique characteristics. Conventionally, EGFR mutations have been classified as sensitive, less sensitive and resistant mutations based on their responses to 1G-TKI. However, recent reports including 2G and 3G-TKI revealed that mutation-specific TKI selection could maximize the benefit for patients with NSCLC harboring less sensitive mutations (Table 4). For further development of targeted therapies with EGFR-TKI, it is important to precisely detect targetable mutations, to select the most appropriate TKI for each mutation and to continue investigating in vitro studies and collecting clinical data for even rare mutations.
Acknowledgments
This work was supported by a Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (16K19989 to Y Kobayashi).
Disclosure Statement
T. Mitsudomi has received lecture fees (from Astra-Zeneca, Boehringer-Ingelheim, and Chugai) and research funding (from Boehringer-Ingelheim and Chugai). Y. Kobayashi has no conflict of interest to declare.